Figure 3.
Differential statistics of immune repertoires across cohorts
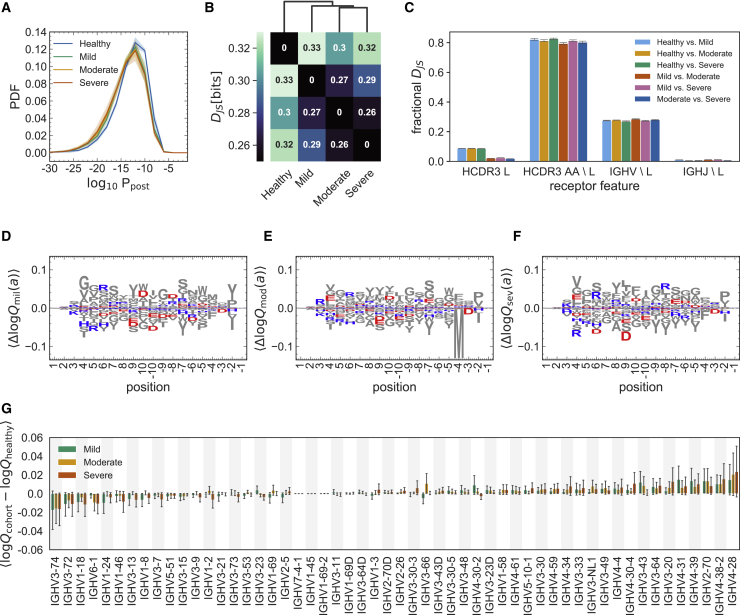
(A) The distribution of the log probability to observe a sequence in the periphery is shown as a normalized probability density function (PDF) for inferred naive progenitors of clonal lineages in cohorts of healthy individuals and the mild, moderate, and severe cohorts of individuals with COVID-19. Full lines show distributions averaged over individuals (biological replicates; Data S1) in each cohort, and shading indicates regions containing one standard deviation of variation among individuals within a cohort.
(B) Clustering of cohorts based on their pairwise Jensen-Shannon divergences (DJS) as a measure of differential selection on cohorts (STAR Methods).
(C) The bar graph shows how incorporating different features into a SONIA selection model contributes to the fractional DJS between models trained on different cohorts. The error bars show the standard deviation of these estimates, using five independent sets of 100,000 generated BCRs for each selection model (STAR Methods).
(D–F) Logo plots show the expected differences in the log-selection factors for amino acid usage, , for the (D) mild, (E) moderate, and (F) severe COVID-19 cohorts. The expectation values are evaluated on the mixture distribution . Positively charged amino acids (lysine, K; arginine, R; and histidine, H) are shown in blue, and negatively charged amino acids (aspartate, D, and glutamate, E) are shown in red. All other amino acids are shown in gray. Positions along the HCDR3 are shown up to 10 residues starting from the 3′ (positive values) and 5′ ends (negative values).
(G) The bar graph shows the average mean difference between the log-selection factors for IGHV gene usage for the mild (green), moderate (yellow), and severe (red) COVID-19 cohorts, with the mean differences computed using the mixture distribution , and the average is taken over the 30 independently trained SONIA models for each cohort. Error bars show standard deviation of these estimates across the inferred SONIA models (STAR Methods).