Abstract
The 5′ cap and 3′ poly(A) tail of mRNA are known to synergistically regulate mRNA translation and stability. Recent computational and experimental studies revealed that both protein-coding and non-coding RNAs will fold with extensive intramolecular secondary structure, which will result in close distances between the sequence ends. This proximity of the ends is a sequence-independent, universal property of most RNAs. Only low-complexity sequences without guanosines are without secondary structure and exhibit end-to-end distances expected for RNA random coils. The innate proximity of RNA ends might have important biological implications that remain unexplored. In particular, the inherent compactness of mRNA might regulate translation initiation by facilitating the formation of protein complexes that bridge mRNA 5′ and 3′ ends. Additionally, the proximity of mRNA ends might mediate coupling of 3′ deadenylation to 5′ end mRNA decay.
This article is categorized under:
RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry
RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems
Translation > Translation Regulation
Keywords: end-to-end distance, RNA secondary structure, translation regulation
1 ∣. INTRODUCTION
The sequence and secondary structures at the 5′ and 3′ termini of RNA play important roles in cellular processes (Chatterjee & Pal, 2009; Genuth & Barna, 2018; Hinnebusch, Ivanov, & Sonenberg, 2016). Both the 5′ and 3′ ends of RNA are recognized by proteins that mediate RNA processing or mRNA translation (Curry, Kotik-Kogan, Conte, & Brick, 2009). In eukaryotes, transcripts produced by RNA polymerase II are modified with a 7-methyl-guanosine cap (m7Gppp cap) at the 5′ end and a poly(A) tail at the 3′ end.
Both the 5′ cap and the 3′ poly(A) protect mRNA from degradation and stimulate translation. Furthermore, the regulation of both mRNA degradation and translation involve interactions of the 5′ cap and 3′ poly(A) tail that are protein mediated. The pervasiveness across evolution of protein bridges spanning the ends of mRNA in translation regulation and decay is surprising because mRNA circularization should incur a substantial entropic cost as compared to a random coil (Yoffe, Prinsen, Gelbart, & Ben-Shaul, 2011). However, new studies discussed below suggest that the expected entropic penalty is mitigated by intramolecular basepairing interactions that provide the energetic drive for compaction. The realization that mRNA ends are intrinsically close may have many important mechanistic and evolutionary implications that await further investigation.
2 ∣. RNA HAS THE INTRINSIC PROPENSITY TO FOLD INTO STRUCTURES THAT BRING THE SEQUENCE ENDS CLOSE
2.1 ∣. The formation of intramolecular RNA secondary structure brings the ends into proximity
It has long been known that nucleotides adjacent to the 5′ end are basepaired with nucleotides adjacent to the 3′ end in a number of non-coding RNA molecules, such as tRNA, 5S rRNA, 23S rRNA and the RNA component of RNase P (Fox & Woese, 1975; Gutell, Gray, & Schnare, 1993; Holley et al., 1965; James, Olsen, Liu, & Pace, 1988). The basepairing between the ends in these RNAs is rather amazing considering significant variations in RNA length, from 76 (tRNA) to 2,900 nucleotides (23S rRNA). As more RNA structures were determined over the years, the list of RNA molecules showing basepairing between the 5′ and 3′ ends continued to grow (Vicens, Kieft, & Rissland, 2018). However, this common feature of many RNA structures attracted little attention from investigators.
Recent computational studies, focusing on RNA secondary structure formation, suggested that the ends of RNA sequences are close in space, regardless of sequence composition and length (Clote, Ponty, & Steyaert, 2012; Fang, 2011; Yoffe et al., 2011). Yoffe et al. (2011) estimated that the average 5′ to 3′ end distance in RNAs is 3 nm. The proximity of RNA ends arises naturally from stem-loop formation. Helices shorten the end-to-end (ETE) distance and, with increasing helix formation, the probability increases that nucleotides at the 5′ end will be basepaired to nucleotides at the 3′ end (Figure 1a,b). It is known from prior studies that even random sequences (composed of all four base identities) will form extensive secondary structure (J. H. Chen, Le, Shapiro, Currey, & Maizel, 1990; Clote, Ferre, Kranakis, & Krizanc, 2005; S. V. Le, Chen, Currey, & Maizel, 1988; S. Y. Le, Chen, & Maizel, 1989; Uzilov, Keegan, & Mathews, 2006; Workman & Krogh, 1999).
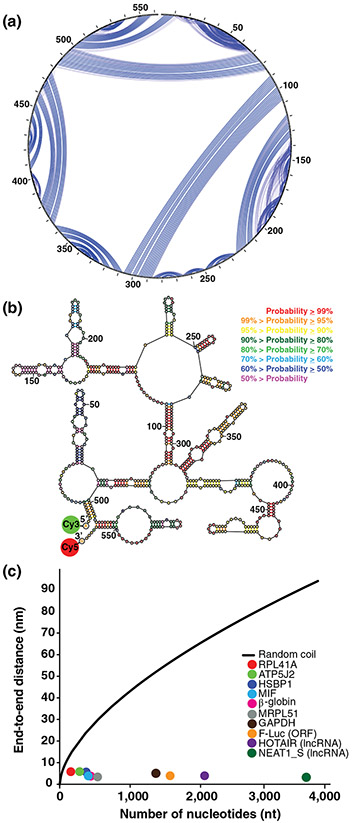
FIGURE 1.
Intramolecular basepairing brings the ends of RNA close. (a) and (b) Show the secondary structure of the human MIF mRNA as predicted and drawn using the RNAstructure software package (https://rna.urmc.rochester.edu/RNAstructure.html). (a) Shows a circular diagram where the sequence is clockwise around the outside of the circle, with the 5′ and 3′ ends at the top of the circle. Blue lines are basepairs; the weight of a blue line represents the estimated pairing probability in the Boltzmann ensemble, where heavier lines are higher estimated probabilities. (b) Shows a collapsed diagram of one secondary structure in the ensemble, where basepairs are colored according to estimated base pairing probabilities in the conformational ensemble. Both representations of the secondary structure demonstrate how basepairing brings the ends close. The probable helix close to the 5′ end and the probable stem-loop at the 3′ end both serve to bring the ends together for this sequence. (c) Shows the FRET-measured end-to-end distances as a function of sequence length. The colored dots are: yeast RPL41A mRNA (red), firefly luciferase mRNA (orange), rabbit β-globin mRNA (magenta), human ATP5J2 mRNA (green), HSBP1 mRNA (indigo), MIF mRNA (blue), MRPL51 mRNA (gray), GAPDH mRNA (brown), HOTAIR lncRNA (purple), and NEAT1_S lncRNA (dark green). The black line is the end-to-end distance of a freely jointed RNA chain (Reprinted with permission from Lai et al. [2018]). FRET, Förster resonance energy transfer
These computational predictions were first tested using several viral RNAs and mRNAs from the fungus Trichoderma atroviride with lengths from 500 to 5,000 nucleotides (Leija-Martinez et al., 2014). These sequences were folded in vitro without any protein factors, and single-molecule Förster resonance energy transfer (smFRET) was observed between the two ends of the molecules. When FRET was detected, the ETE distance ranged from 5 to 9 nm (Leija-Martinez et al., 2014). These results are consistent with the computational studies suggesting that the 5′ and 3′ ends in all RNAs are invariably close and tend to basepair to each other.
More recently, we further tested this hypothesis by measuring FRET between donor and acceptor fluorophores introduced at the 5′ end of 5′ UTR and 3′ end of 3′ UTR, respectively, in eight yeast and human mRNAs (Figure 1b) (Lai et al., 2018). We measured ensemble FRET in doubly-labeled mRNA molecules, folded in vitro in the absence of proteins, and FRET was observed for all eight tested mRNAs. The average ETE distances, determined for each transcript from ensemble FRET data, were between 5 and 7 nm. These distances are independent of sequence length (Figure 1c), and these distances are up to 10 times shorter than those predicted by the freely jointed chain model for RNA random coils (Cantor & Schimmel, 1980; Grosberg & Khokhlov, 1994) (Figure 1c). In addition, FRET between fluorophores attached to the 5′ and 3′ ends was detected in two well-studied long non-coding (lnc)RNAs, HOTAIR and NEAT1_S (Figure 1c), providing additional support for the hypothesis about universal closeness of RNA ends (Lai et al., 2018).
Introduction of unstructured sequences, such as CA repeats, into the 5′ and 3′ UTRs of an mRNA led to disappearance of FRET between fluorophores attached to the 5′ end of the 5′ UTR and 3′ end of the 3′ UTR (Lai et al., 2018). These results indicated that the 5′ and 3′ ends of the wild-type mRNA sequence were brought within FRET distance via the formation of intramolecular basepairing. Additional FRET experiments also showed that the poly(A) tail is not involved in basepairing interactions with the 5′ UTR (Lai et al., 2018).
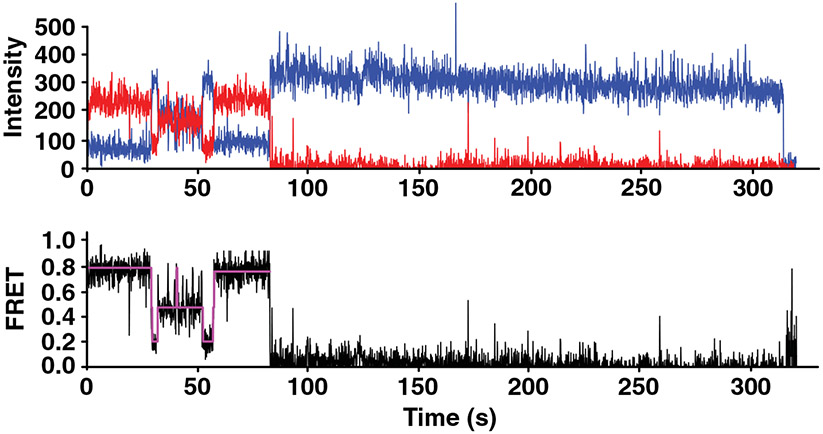
smFRET measurements of individual mRNA molecules revealed that instead of folding into one stable structure, mRNAs fold into an ensemble of several structural states with distinct ETE distances (Figure 2) (Lai et al., 2018) and the strands interconvert between these structures. The spontaneous interconversion, as observed by smFRET traces, demonstrated rates similar to those previously measured for spontaneous transitions between two alternative 5 basepair-long RNA helixes (Furtig et al., 2007) (Figure 2). These data indicated that analogous structural rearrangements spontaneously occur in mRNAs.
FIGURE 2.
smFRET shows that mRNAs fold into a dynamic ensemble of structures. The top plot shows the smFRET for the human GAPDH mRNA as a function of time where blue is fluorescence intensity of the donor (Cy3) at the 3′ end of the 3′ UTR and red is fluorescence intensity of the acceptor (Cy5) at the 5′ end of the 5′ UTR. The acceptor photobleaches at about 80 s. The bottom plot shows the FRET efficiency in black with an idealized model fit by a Hidden Markon Model in magenta, where fluctuation is shown between the 0.2, 0.4, and 0.8 FRET states that correspond to distinct end-to-end distances (Reprinted with permission from Lai et al. [2018]). smFRET, single-molecule Förster resonance energy transfer
2.2 ∣. Most mRNA and lncRNA sequences have the propensity to fold into structures with short ETE distances
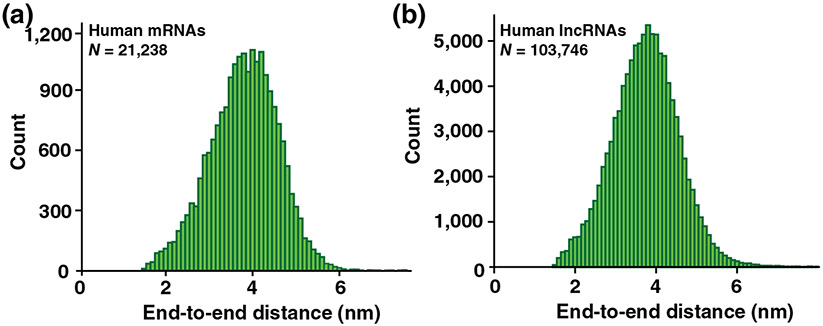
We developed new software within RNAstructure for modeling the distribution of ETE distances of conformational ensembles (Lai et al., 2018). The software utilizes a freely jointed chain polymer theory to estimate the ETE distance for each structure in a stochastically sampled ensemble (Aalberts & Nandagopal, 2010; Ding & Lawrence, 2003; Reuter & Mathews, 2010). The mean ETE distances from this software correlate with our ensemble FRET measurements. We then applied the software to make estimates for the distances between the 5′ end of 5′ UTR and 3′ end of 3′ UTR across the HeLa human cell transcriptome of ~21,000 transcripts. The estimated distances were relatively narrowly distributed with a mean of ~4 nm (Figure 3a). It was rare to have a long ETE distance (defined as longer than 8 nm); about ~0.01% of mRNAs were estimated to have a long ETE distance (Lai et al., 2018). Likewise, the estimated ETE distances in ~104,000 human RNA sequences annotated as lncRNAs were relatively narrowly distributed with a mean of ~4 nm (Figure 3b). Only ~0.12% of all lncRNAs were estimated to have a long ETE distance (Lai et al., 2018). Hence, the intrinsic propensity of RNA structure to result in short ETE distances appear to be common to all human mRNAs and lncRNAs.
FIGURE 3.
Histogram of end-to-end distances for human mRNAs and lncRNAs. (a) Distribution of estimated mRNA end-to-end distances for the HeLa cell transcriptome. (b) Distribution for human lncRNA sequences. N is the number of sequences analyzed (Reprinted with permission from Lai et al. [2018])
Importantly, the proximity of RNA ends appears to be largely independent of sequence and length (Clote et al., 2012; Lai et al., 2018; Yoffe et al., 2011). For example, when 10,000 variants of human GAPDH mRNA were generated computationally by shuffling the 3′ UTR sequence (therefore preserving the original adenosine/guanosine/cytosine/uracil ratio), the estimated ETE distance in these variants was narrowly distributed with a population mean of 4 nm. These calculations were validated by FRET measurements showing that FRET values in the wild-type and a shuffled GAPDH mRNA were indistinguishable. These data support the idea that the closeness of RNA ends is an inherent property of RNA as a polymer rather than a product of natural selection.
2.3 ∣. mRNA 5′ and 3′ UTRs basepair in vivo
The intrinsically close ETE distances in mRNA and lncRNA are likely also found in live cells. mRNA secondary structure might be disrupted by helicases and other RNA binding proteins. Furthermore, the ribosome efficiently unwinds the secondary structure of mRNA within an open reading frame (ORF) by translocating along the mRNA during the elongation of the polypeptide chain (Takyar, Hickerson, & Noller, 2005; Wen et al., 2008). Nevertheless, despite the presence of RNA helicases, a number of structured mRNA elements are known to regulate translation initiation, including bacterial riboswitches (Roth & Breaker, 2009), frameshift-inducing hairpins and pseudoknots of eukaryotic viruses (Giedroc & Cornish, 2009), internal ribosome entry sites (Mauger, Siegfried, & Weeks, 2013), iron response elements in the 5′ UTR of transcripts coding for proteins involved in iron metabolism (Leipuviene & Theil, 2007), and cap-independent translational enhancers (CITEs) (Simon & Miller, 2013). Studies have also shown that protein binding sites on mRNA are determined by accessibility, as governed by the RNA structure (Li, Kazan, Lipshitz, & Morris, 2014; Li, Quon, Lipshitz, & Morris, 2010). Similarly, accessibility to oligonucleotide binding in siRNAs, miRNAs, and antisense oligonucleotides is also governed by the RNA structure, which can occlude sites (Z. J. Lu & Mathews, 2008a, 2008b; Shao et al., 2007; Tafer et al., 2008). Hence, mRNA secondary structure of mRNA is important in the cell in spite of the activities of helicases and other RNA-binding proteins (Aw et al., 2016; Z. Lu et al., 2016; Sharma, Sterne-Weiler, O'Hanlon, & Blencowe, 2016; Wu & Bartel, 2017).
Transcriptome-wide chemical probing studies in yeast, plant and human cells also support the importance of secondary structure in vivo (Aw et al., 2016; Ding et al., 2014; Z. Lu et al., 2016; Rouskin, Zubradt, Washietl, Kellis, & Weissman, 2014; Sharma et al., 2016; Ziv et al., 2018). Intramolecular basepairing between distant segments of mRNA, including interactions between the 5′ and 3′ UTRs, were also observed by mapping of RNA–RNA interactions by psoralen cross-linking in yeast and human cells (Aw et al., 2016; Z. Lu et al., 2016; Sharma et al., 2016; Ziv et al., 2018). These experimental results support the idea that the intrinsic propensity of RNAs to fold into structures with short ETE distances applies to RNA folding within cells.
Several lines of evidence suggest that mRNA secondary structure in the cell is dynamic and undergoes constant rearrangements. For example, transcriptome-wide probing of RNA secondary structure by proximity ligation revealed a prevalence of alternative structures in multiple transcripts (Z. Lu et al., 2016; Ziv et al., 2018). Some of these rearrangements might be triggered by binding and disassociation of RNA binding proteins. For example, single-molecule-resolution fluorescent in situ hybridization (smFISH) and proximity-ligation studies indicated a lack of interactions between distant segments of nuclear mRNAs bound with exon junction protein complexes (EJCs) (Adivarahan et al., 2018; Metkar et al., 2018). By contrast, smFISH demonstrated that the 5′ and 3′ ends of cytoplasmic mRNAs, which are not actively translated, are co-localized (Adivarahan et al., 2018; Khong & Parker, 2018). Translation by the ribosome likely plays a major role in remodeling mRNA structure (Khong & Parker, 2020). The pioneer round of mRNA translation by the ribosome, which displaces EJCs and other proteins deposited on mRNA in the nucleus (Maquat, Tarn, & Isken, 2010), may enable mRNA remodeling and folding into structures with short ETE distances after termination of protein synthesis. By contrast, mRNA translation in poly-ribosomes likely keeps the mRNA ORF devoid of secondary structure due to ribosome helicase activity (Adivarahan et al., 2018; Khong & Parker, 2018, 2020). Hence, it is likely that in vivo, each mRNA folds into structures with short ETE distances only for a fraction of mRNA lifetime.
3 ∣. BIOLOGICAL IMPLICATIONS OF THE INTRINSIC COMPACTNESS OF RNA
3.1 ∣. The intrinsic closeness of RNA ends as an evolutionary hurdle
High basepairing potential and intrinsic compactness of most natural RNA sequences have multiple evolutionary implications. mRNAs and RNA-binding proteins may co-evolve to overcome or, in some cases, to exploit the intrinsic closeness of mRNA ends. Because most, if not all, mRNA and lncRNA sequences have a propensity to form extensive secondary structures, RNA helicases and single-strand RNA binding proteins are required to keep RNA in the single-stranded conformation in live cells. RNA unwinding likely constitutes a significant energy expenditure for the cell. When RNA helicases and single-strand RNA binding proteins disassociate from RNA, RNA likely rapidly folds into compact structures.
The tendency of RNA ends to basepair to each other creates an evolutionary hurdle when RNA function requires one of the RNA ends to form intermolecular baseparing interactions with another RNA molecule or to bind protein factors. One evolutionary strategy to overcome this problem might be favoring intrinsically unstructured sequences at one RNA end. For example, poly(A) sequences in the 5′ UTR of poxvirus mRNAs (Shirokikh & Spirin, 2008) and CAA nucleotide triplet repeats in the Ω leader (5′ UTR) of tobacco mosaic virus mRNAs (Agalarov, Sakharov, Fattakhova, Sogorin, & Spirin, 2014) facilitate the recruitment of the small ribosomal subunit and make translation initiation on these mRNAs independent of the presence of the 5′ cap on mRNA.
What are the defining properties of intrinsically unstructured sequences? To better understand the connection between sequence and ETE distance of RNA, as mediated by basepairing, we developed software to manipulate sequences to adjust ETE distances. We used a genetic algorithm, a type of in silico evolution algorithm (Lai et al., 2018), to randomly evolve a population of sequences that avoid intramolecular basepairing. At each step, either mutation or crossover (a combination of two sequences from the population) occurs, and then sequences with the best fitness are retained for subsequent refinement. Our fitness metric can be tailored to the goal, and the first metric was low mean basepairing probabilities for a stretch of the sequence. In silico evolution of the human GAPDH mRNA sequence confirmed that a reduction in average basepairing probability leads to an increased ETE distance of RNA (Lai et al., 2018). We also observed that in order to reduce basepairing probabilities, an RNA sequence increases the cytosine content and also nearly eliminates guanines. Guanosines form Watson–Crick G-C and also wobble G-U pairs, which have folding stabilities that are similar to Watson–Crick A-U base pairs and are nearly isosteric to A-U pairs (J. L. Chen et al., 2012; Varani & McClain, 2000). Therefore, it appears likely that sequences must avoid guanines to avoid basepairing. As sequences are evolved in silico to reduce the average basepairing probability, we also observe that the linguistic complexity decreases (Gabrielian & Bolshoy, 1999; Troyanskaya, Arbell, Koren, Landau, & Bolshoy, 2002). The complexity is a measure of the sequence repetition, where low complexity indicates that the sequences are repetitive (Lai et al., 2018).
Our in silico evolution experiments showed that guanosine depletion and low sequence complexity are necessary but not sufficient for an RNA to be unstructured (Lai et al., 2018). Only specific, low-complexity sequences of adenosines, cytosines, and uracils will be random coils. Therefore, unstructured RNA sequences in organisms are likely to have resulted from intense natural selection, and theses sequences are likely serving biological roles. Transcriptome-wide search for the intrinsically unstructured RNA segments might reveal novel regulatory sequences in mRNA.
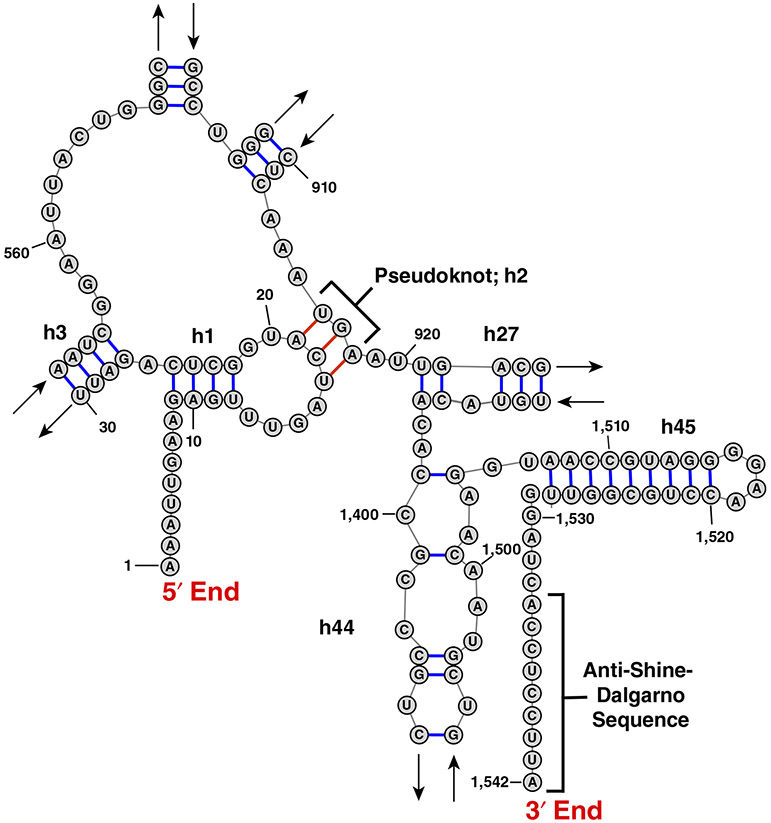
Another evolutionary strategy for keeping RNA ends apart may be the sequestration of one of the RNA ends by pseudoknotted basepairing interactions with nucleotides in the middle of the RNA. 16S rRNA is an example of this strategy. In contrast to 5S or 23S rRNAs, whose ends are basepaired, the 5′ and 3′ ends of 16S rRNA in the small ribosomal subunit are over 8 nm away from each other. Translation initiation in bacteria is facilitated by baseparing interactions between the 3′ end of 16S rRNA and the Shine–Dalgarno (ribosome-binding) sequence in mRNA (Figure 4). Residues adjacent to the 5′ end of 1,542 nucleotide-long 16S rRNA (Escherichia coli numbering) form intramolecular helixes h2 (a pseudoknot) and h3 by basepairing with residues 916–918 and residues 547–556, respectively (Noller & Woese, 1981; Yusupov et al., 2001). These interactions might have evolved to keep the 3′ end of 16S rRNA free to bind the Shine–Dalgarno sequence in mRNAs.
FIGURE 4.
The E. coli 16S rRNA sequence ends are far apart. This figure shows how a pseudoknot in the small subunit rRNA facilitates a longer end-to-end distance than we found in other ncRNA. In part, this exposes the anti-Shine–Dalgarno sequence to base pairs with an mRNA Shine–Dalgarno sequence to initiate translation
3.2 ∣. mRNA compactness in mRNA decay
Closeness of RNA ends may not always be an evolutionary obstacle. Indeed, as discussed below, a number of RNA binding protein complexes emerged through evolution to regulate translation initiation and mRNA decay by bridging mRNA ends. Binding of these complexes to mRNA is likely stabilized by the intrinsic closeness of mRNA ends. The prominence of protein-mediated interactions between mRNA ends in evolution of protein synthesis and mRNA decay suggests that the protein complexes might have evolved to exploit the intrinsic closeness of RNA ends. Therefore, the innate compactness of mRNA likely facilitates the regulation of translation and mRNA degradation.
Protein-mediated interactions between mRNA ends are involved in regulation of mRNA degradation. Eukaryotes have two major pathways of mRNA decay: exosome-catalyzed degradation from the 3′ end and 5′-to-3′ degradation. 5′-to-3′ mRNA degradation begins with poly(A) tail shortening or deadenylation carried out by two multiprotein complexes, PAN2-PAN3 and CCR4-NOT. Dedadenylation is followed by the recruitment of the Dcp1-Dcp2-Edc4 de-capping complex to 5′ end of mRNA, the 5′ cap removal and 5′-to-3′ exonucleolytic degradation of mRNA by Xrn1 exonuclease (Mugridge, Coller, & Gross, 2018). mRNA deadenylation and decapping are coupled through protein–protein interactions of CCR4-NOT and the decapping complex (Mugridge et al., 2018). In budding and fission yeast, deadenylation is also linked to decapping by the interaction between the Pat1-Lsm1-7 complex, which recognizes shortened poly(A) tails, and decapping enzyme Dcp2 (Charenton et al., 2017). Hence, coupling of deadenylation and decapping in 5′-to-3′ mRNA decay involves the formation of protein bridges between mRNA ends. Binding of these protein bridges to mRNA ends is likely stabilized by the intrinsic closeness of mRNA ends.
3.3 ∣. mRNA compactness and the closed-loop model of translation initiation
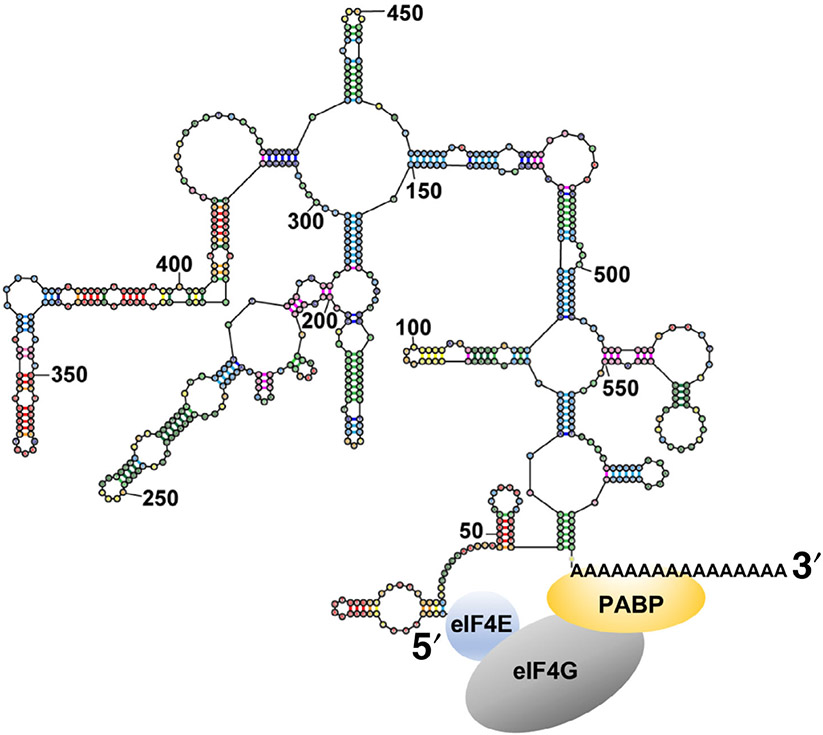
Another example of protein-mediated interactions between the 5′ cap and 3′ poly(A) tail is the formation of a closed-loop structure during translation initiation. To begin protein synthesis, a complex containing the small ribosomal subunit and a number of initiation factors is recruited to the m7Gppp cap structure at the 5′ end of the mRNA (Sonenberg, 2008). The cap structure is recognized by initiation factor eIF4E (eukaryotic Initiation Factor 4E). A complex, called eIF4F, which is formed by eIF4E and two other initiation factors eIF4G and eIF4A, recruits the small ribosomal subunit preassembled with initiator tRNA and initiation factors 1, 1A, 2, 3 and 5 (Aitken & Lorsch, 2012). After recruitment to the 5′ end of mRNA, the small (40S) ribosomal subunit is believed to scan the 5′ UTR of mRNA until it reaches the start codon.
The poly(A) tail at the 3′ end of mRNA was shown to stimulate translation initiation (Jacobson & Favreau, 1983; Munroe & Jacobson, 1990). A number of studies showed that stimulation of translation by the combination of a cap and a poly(A) tail is greater than the product of stimulatory effects of a cap and a poly(A) tail alone (Thompson & Gilbert, 2017). This phenomenon was described as “synergy” between the 5′ cap and 3′ poly(A) tail (Gallie, 1991). The cap-poly(A) tail synergy is believed to be mediated by the binding of cap-binding factor eIF4E and poly(A) binding protein (PABP) to different parts of eIF4G (Figure 5) (Kahvejian, Svitkin, Sukarieh, M'Boutchou, & Sonenberg, 2005; Tarun & Sachs, 1996; Tarun, Wells, Deardorff, & Sachs, 1997). The eIF4E•eIF4G•PABP complex was thought to “circularize” the mRNA (making a “closed loop”) (Jacobson, 1996; Wells, Hillner, Vale, & Sachs, 1998). Mounting evidence suggests that mRNAs vary in the degree to which their translation depends on the presence of eIF4G-PABP interactions (Amrani, Ghosh, Mangus, & Jacobson, 2008; Arava et al., 2003; Archer, Shirokikh, Beilharz, & Preiss, 2016; Thompson & Gilbert, 2017; Thompson, Rojas-Duran, Gangaramani, & Gilbert, 2016). Nevertheless, the interaction between eIF4E, eIF4G and PABP is conserved from yeast to humans and is thought to play an important role in the initiation of protein synthesis in eukaryotes.
FIGURE 5.
The intrinsic closeness of mRNA ends may augment translation by stabilizing the eIF4E•eIF4G•PABP complex
Contrary to the idea of eIF4E•eIF4G•PABP-driven circularization of mRNA, the evidence suggests that in cells, intramolecular basepairing of the mRNA is bringing together the 3′ end of the 3′ UTR and the 5′ end of mRNA rather than the eIF4E•eIF4G•PABP complex. The intrinsic closeness of mRNA ends may arise from basepairing interactions between the 5′ and 3′ UTRs and the formation of stem-loops across the entire mRNA sequence that result in mRNA compactization. One line of evidence is that the eIF4E•eIF4G complex was found to crosslink to the 3′ end the 3′ UTR of yeast transcripts in cells without PABP (Archer, Shirokikh, Hallwirth, Beilharz, & Preiss, 2015). Another line of evidence comes from smFISH imaging studies in human cells. The median distance between the 5′ and 3′ ends of actively translated mRNAs, with lengths from 6,000 to 18,000 nucleotides, was 100–200 nm (Adivarahan et al., 2018; Khong & Parker, 2018). By contrast, the 5′ and 3′ ends in these mRNAs co-localized when translation was inhibited with arsenite or the antibiotic puromycin (Adivarahan et al., 2018; Khong & Parker, 2018). Strikingly, the colocalization of mRNA ends was observed in cells in which the eIF4G•PABP interaction was disrupted by mutagenesis (Adivarahan et al., 2018). Based on these experiments, it was concluded that, in the absence of helicase activity of translating ribosomes, mRNA ends come in close proximity because of intramolecular basepairing interactions within mRNA (Adivarahan et al., 2018; Khong & Parker, 2018). Hence, at least in the translationally repressed state of mRNA, for example, in mRNAs sequestered into stress granules (Adivarahan et al., 2018; Khong & Parker, 2018), mRNA ends are brought in close proximity by mRNA secondary structure. These studies also indicated that the cap•eIF4E•eIF4G•PABP•poly(A) complex is not constitutively bound to actively translated mRNAs. Hence, the cap•eIF4E•eIF4G•PABP•poly(A) interactions may be transient and form during the transition from a translationally inactive to translationally active state of mRNA.
By bringing the 5′ and 3′ ends in the proximity of a few nm, mRNA intramolecular secondary structure decreases the entropic penalty for the cap•eIF4E•eIF4G•PABP•poly(A) complex formation. In other words, PABP binding to the poly(A) tail can facilitate the recruitment of the cap-binding protein complex, eIF4E•eIF4G, to the 5′ end of the mRNA because the 5′ and 3′ ends of mRNA are intrinsically close (Figure 5). Hence, the eIF4G-PABP interaction may have emerged throughout evolution to exploit the intrinsic closeness of mRNA ends.
3.4 ∣. The intrinsic closeness of mRNAs ends may facilitate regulation of translation initiation
While the role of the intrinsic closeness of mRNA ends in the cap•eIF4E•eIF4G•PABP•poly(A) complex formation needs to be further examined, involvement of basepairing interactions between the 5′ and 3′ ends of mRNA in translation initiation of positive-strand RNA plant viruses has been demonstrated experimentally. Translation of these mRNAs, which lack the 5′ cap and poly(A) tail, requires the 3′ CITEs sequence at the 3′ end of viral transcripts (Nicholson & White, 2011). The 3′ CITEs bind eIF4E•eIF4G (or eIF4G alone) and then recruit these initiation factors to the 5′ end of viral transcripts in the process that involves basepairing interactions between the 3′ CITE and the 5′ UTR of the same transcript (Nicholson & White, 2011).
A similar mechanism may underlie translation of histone mRNAs, which also lack the poly(A) tail. During translation initiation on histone mRNAs, the 40S subunit was shown to be initially recruited to specific structural RNA elements within the ORF and at the 3′ end of the mRNA (Cakmakci, Lerner, Wagner, Zheng, & Marzluff, 2008; Martin et al., 2011). The subsequent binding of the 40S to the start codon near the 5′ end of the transcript is likely facilitated by the intrinsic compactness of the mRNA.
The intrinsic compactness of mRNA may also underlie numerous examples of message-specific translational control of protein expression mediated by the formation of protein bridges between the 5′ cap and specific regulatory sequences in the 3′ UTR. For example, translation of mRNAs containing a cytoplasmic polyadenylation element (CPE) in the 3′ UTR is repressed by recruitment the CPEB•Maskin protein complex to CPE. CPEB•Maskin binds to the eIF4E•5′ cap complex and displaces eIF4G (Sonenberg & Hinnebusch, 2009). Similarly, translation of Drosophila oskar mRNA is inhibited by the binding of the eIF4E•5′ cap to the Bruno•Cup protein complex tethered to the Bruno response element in the 3′ UTR of oskar mRNA (Nakamura, Sato, & Hanyu-Nakamura, 2004). Translational repression of ceruloplasmin mRNA upon interferon-γ treatment is mediated by formation of the GAIT complex assembled from Glu-Pro-tRNA synthetase, NS-associated protein 1, GAPDH, and 60S ribosomal protein L13a, which is released from the 60S subunit by phosphorylation. The GAIT complex simultaneously binds to the GAIT element in the 3′ UTR and eIF4G at the 5′ end of ceruloplasmin mRNA to block recruitment of the small ribosomal subunit (Arif et al., 2018). In all these examples, the intrinsic mRNA compactness may stabilize the protein regulatory complexes.
4 ∣. CONCLUSIONS
Short ETE distances might facilitate the binding of protein factors that regulate translation initiation and mRNA decay by bridging mRNA 5′ and 3′ ends. There has been a long-standing view that protein–protein interactions bring the two ends of mRNAs in proximity. This review focuses on the recent understanding that RNA sequences fold by basepairing to bring the 5′ and 3′ ends in proximity, based on computation and on FRET measurements. Closeness of RNA ends is a property of most, if not all, mRNAs and lncRNAs. Only sequences that are devoid of guanosines and have low sequence complexity are intrinsically unstructured. Alternatively, RNA ends can be kept apart by pseudoknotted basepairing interactions with nucleotides in the middle of the RNA. For example, a pseudoknot in the small subunit rRNA facilitates the formation of a structure that makes the 5′ and 3′ ends distant and also exposes the anti-Shine–Dalgarno sequence for basepairing.
Given the intrinsic propensity of mRNA to fold into structures with short ETE distances, it makes sense that proteins would have evolved to complexes that specifically recognize both ends of mRNA. The prevalence of these protein bridges between the 5′ cap and 3′ UTR in the evolution of translational control supports the hypothesis that the regulatory protein complexes emerged throughout evolution to utilize the intrinsic compactness of mRNA.
BOX 1. IS SHORT ETE DISTANCE A COMMON FEATURE OF RNA AND PROTEINS?
“Closeness of the ends” has somewhat different meaning in the case of proteins and RNAs. We and others compare the ETE distance in folded RNA with the ETE distance expected for the random coil conformation of RNA. In this context, the closeness of mRNA (or lncRNA) ends means that the distance between the 5′ and 3′ ends is substantially shorter than the distance expected for the random coil conformation of the sequence. With the exception of few RNAs, including 16S rRNA, 23S rRNA, some RNA aptamers and some ribozymes, most of RNA molecules including mRNAs do not fold into highly condensed structures (Seetin & Mathews, 2011; Yoffe et al., 2008). Because of this, closeness of RNA ends is counterintuitive and is rather remarkable.
In contrast to RNA, most proteins fold into compact, globular structures. For that reason, the distance between N and C termini is usually examined in regard to dimensions of folded proteins (e.g., radius of gyration). In this context, the closeness of protein termini implies that the distance between N and C termini is significantly shorter than the distance expected based on chance and dimensions of a given protein. There is no agreement between different analyses on whether protein termini are generally closer than expected by chance or not (Christopher & Baldwin, 1996; Thornton & Sibanda, 1983). Nevertheless, similar to RNA, in most proteins, ETE distance is shorter in the folded state than in unfolded ensembles of proteins (Schuler & Eaton, 2008)
BOX 2. COMPUTATIONAL TOOLS FOR MANIPULATING RNA BASEPAIRING PROBABILITIES AND ETE DISTANCES.
To evolve sequences to have a longer ETE distance, we wrote the orega (“optimize RNA ends with a genetic algorithm”) program. This is part of the RNAstructure software package, available at https://rna.urmc.rochester.edu. Orega uses a genetic algorithm to randomly evolve a region of an input sequence to maximize its fitness. Fitness is defined as the sum of the mean probability that nucleotides are unpaired in the region, as estimated by secondary structure prediction (Mathews, 2004), and the linguistic complexity in the region. The linguistic complexity quantifies the sequence diversity, where larger values (bounded by 1) indicate that the sequence has little repetition, and small values (bounded by 0) indicate sequence repeats (Gabrielian & Bolshoy, 1999; Trifonov, 1990). We found the complexity was an important aspect to evolve sequences that could be successfully cloned
The other software tool we developed is the ETEcalculator program, which estimates the ETE distance for an input RNA sequence. ETEcalculator samples structures from the Boltzmann ensemble (Ding & Lawrence, 2003) and estimates the ETE distance for each structure using polymer theory (Aalberts & Nandagopal, 2010). The estimated ETE distance is the mean distance across the sampled structures as reported in nm.
ACKNOWLEDGMENTS
This work was supported in the Ermolenko and Mathews laboratories by NIH grants R01GM132041 (to D. N. E.) and R01GM076485 (to D. H. M.).
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
REFERENCES
- Aalberts DP, & Nandagopal N (2010). A two-length-scale polymer theory for RNA loop free energies and helix stacking. RNA, 16(7), 1350–1355. 10.1261/rna.1831710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adivarahan S, Livingston N, Nicholson B, Rahman S, Wu B, Rissland OS, & Zenklusen D (2018). Spatial organization of single mRNPs at different stages of the gene expression pathway. Molecular Cell, 72(4), 727–738 e725. 10.1016/j.molcel.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalarov S, Sakharov PA, Fattakhova D, Sogorin EA, & Spirin AS (2014). Internal translation initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic ribosomal particles. Scientific Reports, 4, 4438. 10.1038/srep04438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, & Lorsch JR (2012). A mechanistic overview of translation initiation in eukaryotes. Nature Structural & Molecular Biology, 19(6), 568–576. 10.1038/nsmb.2303 [DOI] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, & Jacobson A (2008). Translation factors promote the formation of two states of the closed-loop mRNP. Nature, 453(7199), 1276–1280. 10.1038/nature06974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, & Herschlag D (2003). Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 100(7), 3889–3894. 10.1073/pnas.0635171100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, & Preiss T (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature, 535(7613), 570–574. 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Hallwirth CV, Beilharz TH, & Preiss T (2015). Probing the closed-loop model of mRNA translation in living cells. RNA Biology, 12(3), 248–254. 10.1080/15476286.2015.1017242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif A, Yao P, Terenzi F, Jia J, Ray PS, & Fox PL (2018). The GAIT translational control system. Wiley Interdisciplinary Reviews: RNA, 9(2), e1441. 10.1002/wrna.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw JG, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, … Wan Y (2016). In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Molecular Cell, 62(4), 603–617. 10.1016/j.molcel.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, & Marzluff WF (2008). SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Molecular and Cellular Biology, 28(3), 1182–1194. 10.1128/MCB.01500-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor CR, & Schimmel PR (1980). Biophysical chemistry: Part I: The conformation of biological macromolecules (Vol. 1). New York: W.H. Freeman and Company. [Google Scholar]
- Charenton C, Gaudon-Plesse C, Fourati Z, Taverniti V, Back R, Kolesnikova O, … Graille M (2017). A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5′-3′ mRNA exonuclease in yeast. Proceedings of the National Academy of Sciences of the United States of America, 114(45), E9493–E9501. 10.1073/pnas.1711680114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, & Pal JK (2009). Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biology of the Cell, 101(5), 251–262. 10.1042/BC20080104 [DOI] [PubMed] [Google Scholar]
- Chen JH, Le SY, Shapiro B, Currey KM, & Maizel JV (1990). A computational procedure for assessing the significance of RNA secondary structure. Computer Applications in the Biosciences, 6(1), 7–18. 10.1093/bioinformatics/6.1.7 [DOI] [PubMed] [Google Scholar]
- Chen JL, Dishler AL, Kennedy SD, Yildirim I, Liu B, Turner DH, & Serra MJ (2012). Testing the nearest neighbor model for canonical RNA base pairs: Revision of GU parameters. Biochemistry, 51(16), 3508–3522. 10.1021/bi3002709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher JA, & Baldwin TO (1996). Implications of N and C-terminal proximity for protein folding. Journal of Molecular Biology, 257 (1), 175–187. 10.1006/jmbi.1996.0154 [DOI] [PubMed] [Google Scholar]
- Clote P, Ferre F, Kranakis E, & Krizanc D (2005). Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA, 11(5), 578–591. 10.1261/rna.7220505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clote P, Ponty Y, & Steyaert JM (2012). Expected distance between terminal nucleotides of RNA secondary structures. Journal of Mathematical Biology, 65(3), 581–599. 10.1007/s00285-011-0467-8 [DOI] [PubMed] [Google Scholar]
- Curry S, Kotik-Kogan O, Conte MR, & Brick P (2009). Getting to the end of RNA: Structural analysis of protein recognition of 5′ and 3′ termini. Biochimica et Biophysica Acta, 1789(9–10), 653–666. 10.1016/j.bbagrm.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Ding Y, & Lawrence CE (2003). A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Research, 31(24), 7280–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, & Assmann SM (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature, 505(7485), 696–700. 10.1038/nature12756 [DOI] [PubMed] [Google Scholar]
- Fang LT (2011). The end-to-end distance of RNA as a randomly self-paired polymer. Journal of Theoretical Biology, 280(1), 101–107. 10.1016/j.jtbi.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Fox GE, & Woese CR (1975). 5S RNA secondary structure. Nature, 256(5517), 505–507. 10.1038/256505a0 [DOI] [PubMed] [Google Scholar]
- Furtig B, Wenter P, Reymond L, Richter C, Pitsch S, & Schwalbe H (2007). Conformational dynamics of bistable RNAs studied by time-resolved NMR spectroscopy. Journal of the American Chemical Society, 129(51), 16222–16229. 10.1021/ja076739r [DOI] [PubMed] [Google Scholar]
- Gabrielian A, & Bolshoy A (1999). Sequence complexity and DNA curvature. Computers & Chemistry, 23(3–4), 263–274. [DOI] [PubMed] [Google Scholar]
- Gallie DR (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & Development, 5(11), 2108–2116. [DOI] [PubMed] [Google Scholar]
- Genuth NR, & Barna M (2018). Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nature Reviews. Genetics, 19(7), 431–452. 10.1038/s41576-018-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc DP, & Cornish PV (2009). Frameshifting RNA pseudoknots: Structure and mechanism. Virus Research, 139(2), 193–208. 10.1016/j.virusres.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg AY, & Khokhlov AR (1994). Statistical physics of macromolecules. Woodbury, New York: AIP Press. [Google Scholar]
- Gutell RR, Gray MW, & Schnare MN (1993). A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Research, 21(13), 3055–3074. 10.1093/nar/21.13.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, & Sonenberg N (2016). Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science, 352 (6292), 1413–1416. 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, … Zamir A (1965). Structure of a ribonucleic acid. Science, 147(3664), 1462–1465. 10.1126/science.147.3664.1462 [DOI] [PubMed] [Google Scholar]
- Jacobson A (1996). Poly(A) metabolism and tranlsation: The closed-loop model. Translational control (pp. 451–480). New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Jacobson A, & Favreau M (1983). Possible involvement of poly(A) in protein synthesis. Nucleic Acids Research, 11(18), 6353–6368. 10.1093/nar/11.18.6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James BD, Olsen GJ, Liu JS, & Pace NR (1988). The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell, 52(1), 19–26. 10.1016/0092-8674(88)90527-2 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, & Sonenberg N (2005). Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Development, 19(1), 104–113. 10.1101/gad.1262905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, & Parker R (2018). mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. The Journal of Cell Biology, 217(12), 4124–4140. 10.1083/jcb.201806183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, & Parker R (2020). The landscape of eukaryotic mRNPs. RNA, 26(3), 229–239. 10.1261/rna.073601.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WC, Kayedkhordeh M, Cornell EV, Farah E, Bellaousov S, Rietmeijer R, … Ermolenko DN (2018). mRNAs and lncRNAs intrinsically form secondary structures with short end-to-end distances. Nature Communications, 9(1), 4328. 10.1038/s41467-018-06792-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SV, Chen JH, Currey KM, & Maizel JV Jr. (1988). A program for predicting significant RNA secondary structures. Computer Applications in the Biosciences, 4(1), 153–159. 10.1093/bioinformatics/4.1.153 [DOI] [PubMed] [Google Scholar]
- Le SY, Chen JH, & Maizel JV (1989). Thermodynamic stability and statistical significance of potential stem-loop structures situated at the frameshift sites of retroviruses. Nucleic Acids Research, 17(15), 6143–6152. 10.1093/nar/17.15.6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leija-Martinez N, Casas-Flores S, Cadena-Nava RD, Roca JA, Mendez-Cabanas JA, Gomez E, & Ruiz-Garcia J (2014). The separation between the 5′-3′ ends in long RNA molecules is short and nearly constant. Nucleic Acids Research, 42(22), 13963–13968. 10.1093/nar/gku1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipuviene R, & Theil EC (2007). The family of iron responsive RNA structures regulated by changes in cellular iron and oxygen. Cellular and Molecular Life Sciences, 64(22), 2945–2955. 10.1007/s00018-007-7198-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kazan H, Lipshitz HD, & Morris QD (2014). Finding the target sites of RNA-binding proteins. Wiley Interdisciplinary Reviews RNA, 5(1), 111–130. 10.1002/wrna.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, & Morris Q (2010). Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA, 16(6), 1096–1107. 10.1261/rna.2017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, … Chang HY (2016). RNA duplex map in living cells reveals higher-order transcriptome structure. Cell, 165(5), 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, & Mathews DH (2008a). Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Research, 36(2), 640–647. 10.1093/nar/gkm920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, & Mathews DH (2008b). Fundamental differences in the equilibrium considerations for siRNA and antisense oligodeoxynucleotide design. Nucleic Acids Research, 36(11), 3738–3745. 10.1093/nar/gkn266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, & Isken O (2010). The pioneer round of translation: Features and functions. Cell, 142(3), 368–374. 10.1016/j.cell.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, & Eriani G (2011). Cap-assisted internal initiation of translation of histone H4. Molecular Cell, 41(2), 197–209. 10.1016/j.molcel.2010.12.019 [DOI] [PubMed] [Google Scholar]
- Mathews DH (2004). Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA, 10(8), 1178–1190. 10.1261/rna.7650904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger DM, Siegfried NA, & Weeks KM (2013). The genetic code as expressed through relationships between mRNA structure and protein function. FEBS Letters, 587(8), 1180–1188. 10.1016/j.febslet.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar M, Ozadam H, Lajoie BR, Imakaev M, Mirny LA, Dekker J, & Moore MJ (2018). Higher-order organization principles of pre-translational mRNPs. Molecular Cell, 72(4), 715–726 e713. 10.1016/j.molcel.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge JS, Coller J, & Gross JD (2018). Structural and molecular mechanisms for the control of eukaryotic 5′-3′ mRNA decay. Nature Structural & Molecular Biology, 25(12), 1077–1085. 10.1038/s41594-018-0164-z [DOI] [PubMed] [Google Scholar]
- Munroe D, & Jacobson A (1990). mRNA poly(A) tail, a 3′ enhancer of translational initiation. Molecular and Cellular Biology, 10(7), 3441–3455. 10.1128/mcb.10.7.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Sato K, & Hanyu-Nakamura K (2004). Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Developmental Cell, 6(1), 69–78. 10.1016/s1534-5807(03)00400-3 [DOI] [PubMed] [Google Scholar]
- Nicholson BL, & White KA (2011). 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Current Opinion in Virology, 1(5), 373–380. 10.1016/j.coviro.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Noller HF, & Woese CR (1981). Secondary structure of 16S ribosomal RNA. Science, 212(4493), 403–411. 10.1126/science.6163215 [DOI] [PubMed] [Google Scholar]
- Reuter JS, & Mathews DH (2010). RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, & Breaker RR (2009). The structural and functional diversity of metabolite-binding riboswitches. Annual Review of Biochemistry, 78, 305–334. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, & Weissman JS (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature, 505(7485), 701–705. 10.1038/nature12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler B, & Eaton WA (2008). Protein folding studied by single-molecule FRET. Current Opinion in Structural Biology, 18(1), 16–26. 10.1016/j.sbi.2007.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetin MG & Mathews DH (2011). Automated RNA tertiary structure prediction from secondary structure and low-resolution restraints. Journal of Computational Chemistry, 32, 2232–2244. 10.1002/jcc.21806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, & Ding Y (2007). Effect of target secondary structure on RNAi efficiency. RNA, 13(10), 1631–1640. 10.1261/rna.546207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O'Hanlon D, & Blencowe BJ (2016). Global mapping of human RNA-RNA interactions. Molecular Cell, 62 (4), 618–626. 10.1016/j.molcel.2016.04.030 [DOI] [PubMed] [Google Scholar]
- Shirokikh NE, & Spirin AS (2008). Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proceedings of the National Academy of Sciences of the United States of America, 105(31), 10738–10743. 10.1073/pnas.0804940105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, & Miller WA (2013). 3′ Cap-independent translation enhancers of plant viruses. Annual Review of Microbiology, 67, 21–42. 10.1146/annurev-micro-092412-155609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N (2008). eIF4E, the mRNA cap-binding protein: From basic discovery to translational research. Biochemistry and Cell Biology, 86(2), 178–183. 10.1139/O08-034 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, & Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell, 136 (4), 731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, & Hofacker IL (2008). The impact of target site accessibility on the design of effective siRNAs. Nature Biotechnology, 26(5), 578–583. 10.1038/nbt1404 [DOI] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, & Noller HF (2005). mRNA helicase activity of the ribosome. Cell, 120(1), 49–58. 10.1016/j.cell.2004.11.042 [DOI] [PubMed] [Google Scholar]
- Tarun SZ Jr., & Sachs AB (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. The EMBO Journal, 15(24), 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr., Wells SE, Deardorff JA, & Sachs AB (1997). Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proceedings of the National Academy of Sciences of the United States of America, 94(17), 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, & Gilbert WV (2017). mRNA length-sensing in eukaryotic translation: Reconsidering the "closed loop" and its implications for translational control. Current Genetics, 63(4), 613–620. 10.1007/s00294-016-0674-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, & Gilbert WV (2016). The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife, 5, e11154. 10.7554/eLife.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JM, & Sibanda BL (1983). Amino and carboxy-terminal regions in globular proteins. Journal of Molecular Biology, 167(2), 443–460. 10.1016/s0022-2836(83)80344-1 [DOI] [PubMed] [Google Scholar]
- Trifonov EN (1990). Making sense of the human genome. In Sarma RH & Sarma MH (Eds.), Structure and methods (Vol. 1, pp. 69–77). Schenectady, New York: Adenine Press. [Google Scholar]
- Troyanskaya OG, Arbell O, Koren Y, Landau GM, & Bolshoy A (2002). Sequence complexity profiles of prokaryotic genomic sequences: A fast algorithm for calculating linguistic complexity. Bioinformatics, 18(5), 679–688. [DOI] [PubMed] [Google Scholar]
- Uzilov AV, Keegan JM, & Mathews DH (2006). Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics, 7, 173. 10.1186/1471-2105-7-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, & McClain WH (2000). The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Reports, 1(1), 18–23. 10.1093/embo-reports/kvd001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Kieft JS, & Rissland OS (2018). Revisiting the closed-loop model and the nature of mRNA 5′-3′ communication. Molecular Cell, 72(5), 805–812. 10.1016/j.molcel.2018.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, & Sachs AB (1998). Circularization of mRNA by eukaryotic translation initiation factors. Molecular Cell, 2(1), 135–140 doi:S1097-2765(00)80122-7 [pii]. [DOI] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, … Tinoco I (2008). Following translation by single ribosomes one codon at a time. Nature, 452(7187), 598–603. 10.1038/nature06716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, & Krogh A (1999). No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Research, 27(24), 4816–4822. 10.1093/nar/27.24.4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, & Bartel DP (2017). Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell, 169(5), 905–917 e911. 10.1016/j.cell.2017.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe AM, Prinsen P, Gelbart WM, & Ben-Shaul A (2011). The ends of a large RNA molecule are necessarily close. Nucleic Acids Research, 39(1), 292–299. 10.1093/nar/gkq642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, & Ben-Shaul A (2008) Predicting the sizes of large RNA molecules. PNAS October 21, 105(42), 16153–16158. 10.1073/pnas.0808089105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, & Noller HF (2001). Crystal structure of the ribosome at 5.5 A resolution. Science, 292(5518), 883–896. 10.1126/science.1060089 [DOI] [PubMed] [Google Scholar]
- Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, … Miska EA (2018). COMRADES determines in vivo RNA structures and interactions. Nature Methods, 15(10), 785–788. 10.1038/s41592-018-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]