FIGURE 1.
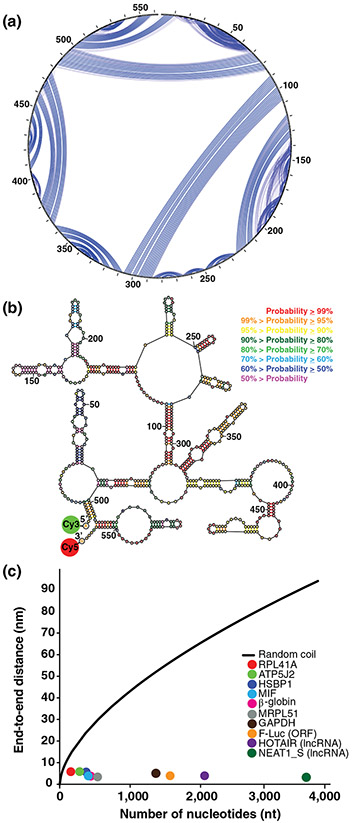
Intramolecular basepairing brings the ends of RNA close. (a) and (b) Show the secondary structure of the human MIF mRNA as predicted and drawn using the RNAstructure software package (https://rna.urmc.rochester.edu/RNAstructure.html). (a) Shows a circular diagram where the sequence is clockwise around the outside of the circle, with the 5′ and 3′ ends at the top of the circle. Blue lines are basepairs; the weight of a blue line represents the estimated pairing probability in the Boltzmann ensemble, where heavier lines are higher estimated probabilities. (b) Shows a collapsed diagram of one secondary structure in the ensemble, where basepairs are colored according to estimated base pairing probabilities in the conformational ensemble. Both representations of the secondary structure demonstrate how basepairing brings the ends close. The probable helix close to the 5′ end and the probable stem-loop at the 3′ end both serve to bring the ends together for this sequence. (c) Shows the FRET-measured end-to-end distances as a function of sequence length. The colored dots are: yeast RPL41A mRNA (red), firefly luciferase mRNA (orange), rabbit β-globin mRNA (magenta), human ATP5J2 mRNA (green), HSBP1 mRNA (indigo), MIF mRNA (blue), MRPL51 mRNA (gray), GAPDH mRNA (brown), HOTAIR lncRNA (purple), and NEAT1_S lncRNA (dark green). The black line is the end-to-end distance of a freely jointed RNA chain (Reprinted with permission from Lai et al. [2018]). FRET, Förster resonance energy transfer