FIGURE 3.
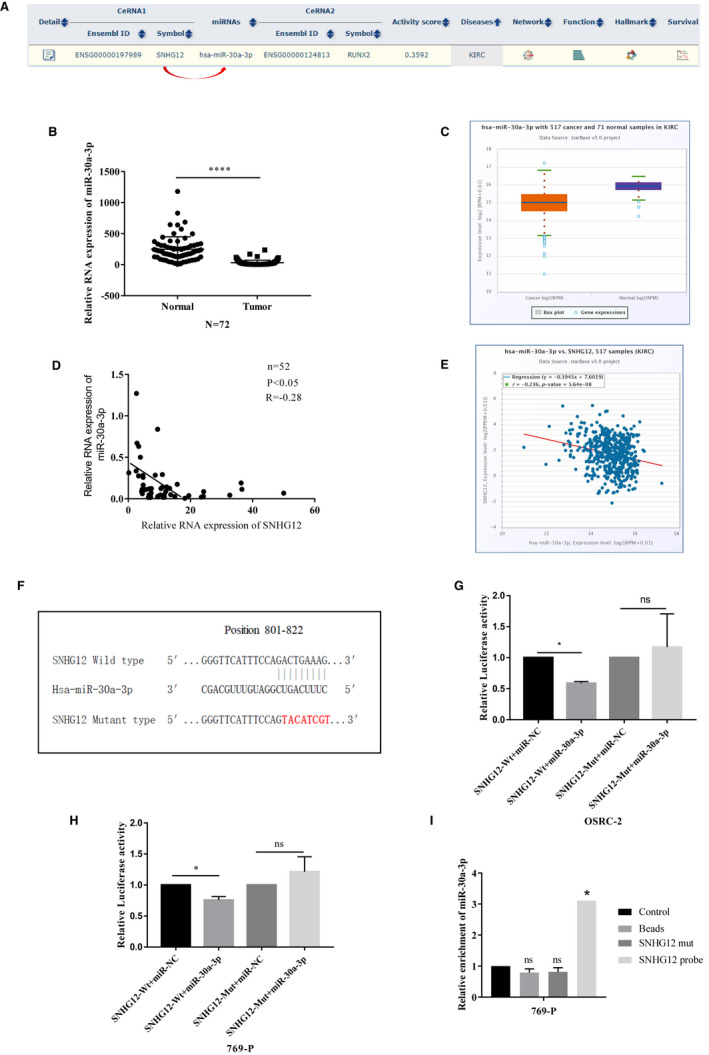
SNHG12 directly interacts with has‐miR‐30a‐3p gene in ccRCC cells. A, Bioinformatic software (ENCORI) suggested that SNHG12 could act as a ceRNA and interacted with has‐miR‐30a‐3p in kidney clear cell carcinoma data set. B, miR‐30a = 3p is significantly down‐regulated in ccRCC tumour tissues compared with adjacent normal tissues. (n = 72, P < .001). C, Data from TCGA database suggested that expression of miR‐30a‐3p in ccRCC tumour tissues (n = 523) is much lower than that in normal kidney tissues (n = 100, P < .001). D, Two‐tailed Pearson's correlation coefficient test suggested a positive correlation between expression of SNHG12 and miR‐30a‐3p in 52 ccRCC tumour tissues (P < .05, r = −.28). E, Data from TCGA database suggested a positive correlation between expression of SNHG12 and miR‐30a‐3p in 517 ccRCC tumour tissues (P < .01, r = −.236). F, Diagram shows the wild‐type and mutant type of miRNA response elements within SNHG12 gene. G and H, Dual luciferase assay suggested that miR‐30a‐3p significantly suppressed the transcriptional level of luciferase vector harbouring wild‐type SNHG12 gene, whereas had no effects on the transcription of luciferase vector harbouring mutant‐type SNHG12 gene (P < .05, ns: not significant). I, RNA pull‐down assay was performed to measure the interaction between SNHG12 and miR‐30a‐3p. miR‐30a‐3p was enriched in biotinylated SNHG12 anti‐sense oligo pull‐down product (P < .05, ns: not significant)
