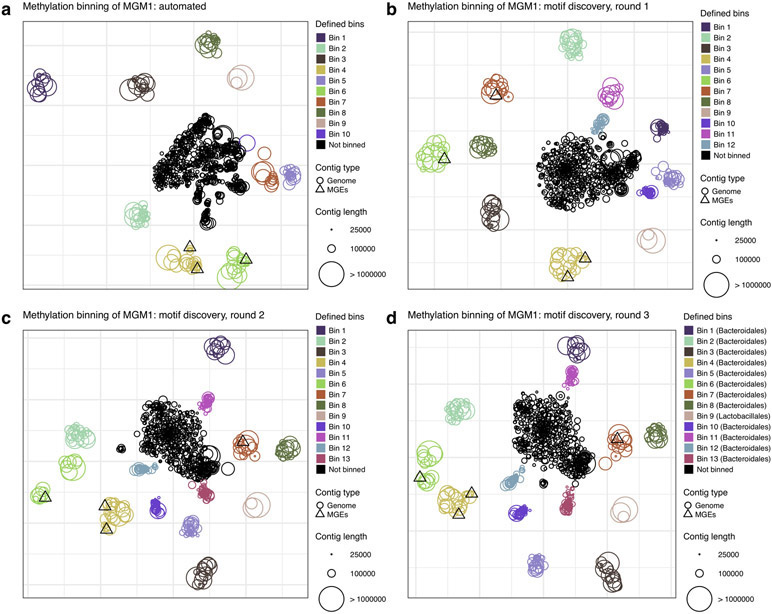
Extended Data Fig. 9. Detailed methylation analysis of MGM1 sample.
(a) Methylation binning using automated methylation features selection (without precise methylation motif discovery; Methods). Methylation features are projected on two dimensions using t-SNE. Contigs are colored per bin defined using DBSCAN, with point sizes matching contig length according to the legend. Two bins with the same methylation motifs were manually merged into Bin 4. (b) Methylation binning using de novo discovered motifs on each bin found in a (Methods). Methylation features computed from de novo discovered motifs are projected on two dimensions using t-SNE. Contigs are colored per bin defined using DBSCAN except Bin 11, which was manually defined. (c) Methylation binning using de novo discovered motifs on each bin found in b. Contigs are colored per bin defined using DBSCAN except for Bin 13, which was manually defined. (d) Methylation binning of MGM1 metagenome contigs using de novo discovered motifs (after three rounds of motif discovery (same as Fig. 5a).