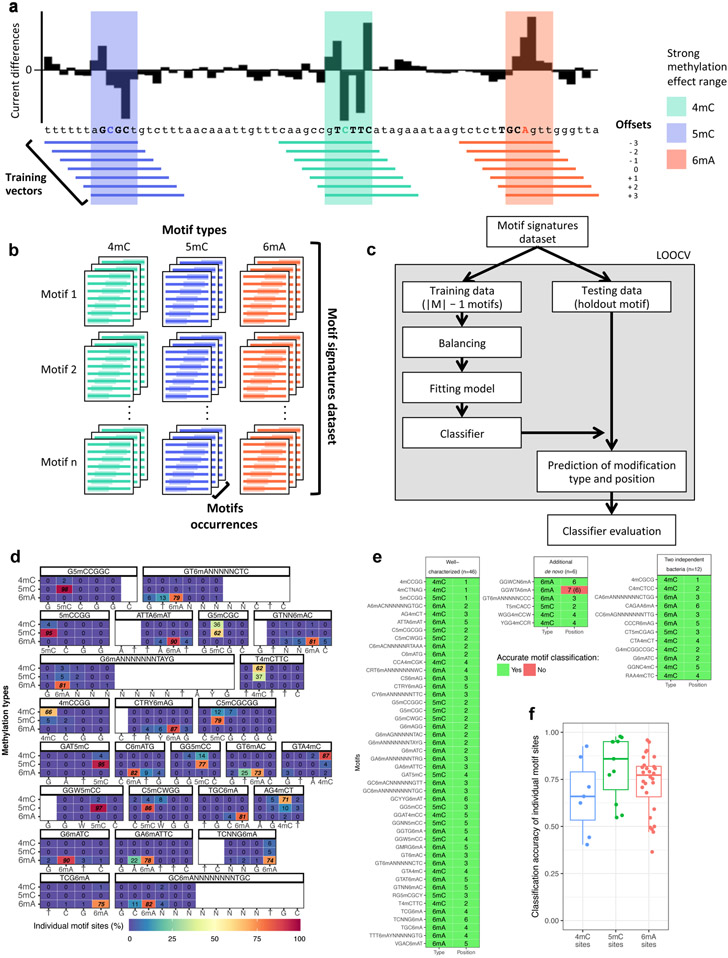
Figure 4:
Classification and fine mapping of three types of DNA methylation. (a) For each motif occurrence, we produced 7 training vectors of length 12 with +/− offsets from 0 to 3 position(s) relative to current differences core defined as [−2, +3] (Extended Data Fig. 1a-c). (b) Each training vector is labeled with methylation type and offset used. They are gathered into a training dataset of current differences flanking 183,818 methylated bases from 46 distinct motifs (Methods). (c) Description of the classifier performance evaluation using leave-one-out cross-validation (LOOCV). (d) Detailed classifier evaluation results for neural network model from the LOOCV evaluation for a subset of the 46 well-characterized methylation motifs are displayed for illustration. Filling colors correspond to percentage of occurrences classified to a specific class: blue (0%) to red (100%). Prediction percentages of expected classes are displayed in italics and fine mapped methylated positions in each motif are displayed in bold. (e) Summary of methylation motifs typing and fine mapping results from the neural network model. Green shows accurately typed and/or fine mapped methylation in motif, while red shows inaccurate prediction with the expected result in parentheses. LOOCV results are used for the “Well-characterized” motifs (n=46), while classification results from the final neural network model trained on the 46 well-characterized motifs are used for both “Additional de novo (n=6)” and “Two independent bacteria (n=12)” motifs. (f) Classification accuracy for individual motifs sites (n=46 motifs including 6mA: 28, 4mC: 7, 5mC: 11) from the neural network model. The lower and upper hinges correspond to the 25th and 75th percentiles while the lower and upper whisker extends to the minima and maxima respectively (capped at 1.5 time the inter-quartile range).