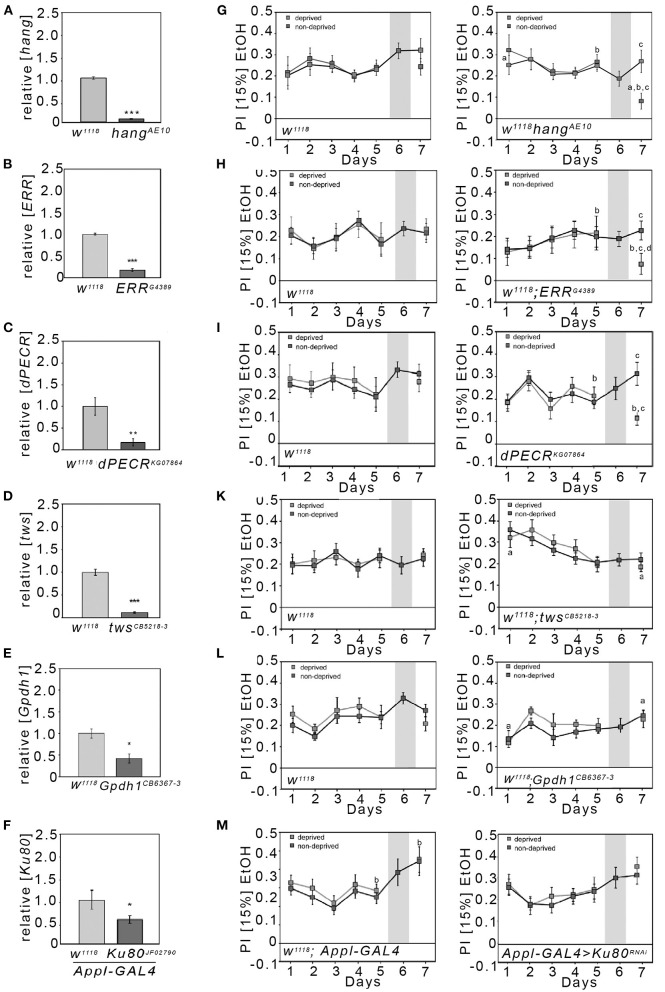
Figure 2.
Reduction in the preference to consume EtOH after abstinence in hang, ERR, and dPECR mutants. (A–F) The qRT-PCR analysis performed on polyA-selected head RNA revealed a significant reduction in transcripts of candidate genes (transcript levels for hangAE10: 0.08 ± 0.01; for ERRG4389: 0.17 ± 0.02; dPECRKG07864: 0.17 ± 0.04; twinsCB5218−3: 0.11 ± 0.02; Gpdh1CB6367−3: 0.42 ± 0.11; for transheterozygous flies carrying a Ku80 RNAi transgene under the control of the Appl-Gal4 driver: 0.55 ± 0.09). N = 4 different RNA samples; data are presented as the means ± STDEV. Significance was determined using a two-tailed Student's t test. *P < 0.05, **P < 0.01, and ***P < 0.001. (G–M) Two different groups of flies were tested—the respective control for the mutants (G–M first panel) and the mutants (G–M second panel). The control group and mutant groups were further divided into two groups. The control group continuously had access to both solutions from days 1 to 7, whereas the experimental group on day 6 had only access to food without ethanol and on day 7 access to both solutions again. (M) Heterozygous Appl-Gal4 flies were compared with the transheterozygous flies, which were carrying one copy of the RNAi transgene and one copy of the Appl-Gal4 transgene. (G–M) The letter “a” indicates a significant difference between the ethanol consumption preference on days 1 and 7; the letter “b” indicates a difference between day 5 prior to the deprivation of ethanol and day 7; and the letter “c” indicates the significant difference between the control group and deprived group at day 7. For a–c, P < 0.016. N = 8–17. The data are presented as the means ± s.e.m. Significance was determined using a two-tailed t tests. To determine whether flies preferred ethanol and did not choose at random between the two offered solutions, the one-sample sign tests were used.