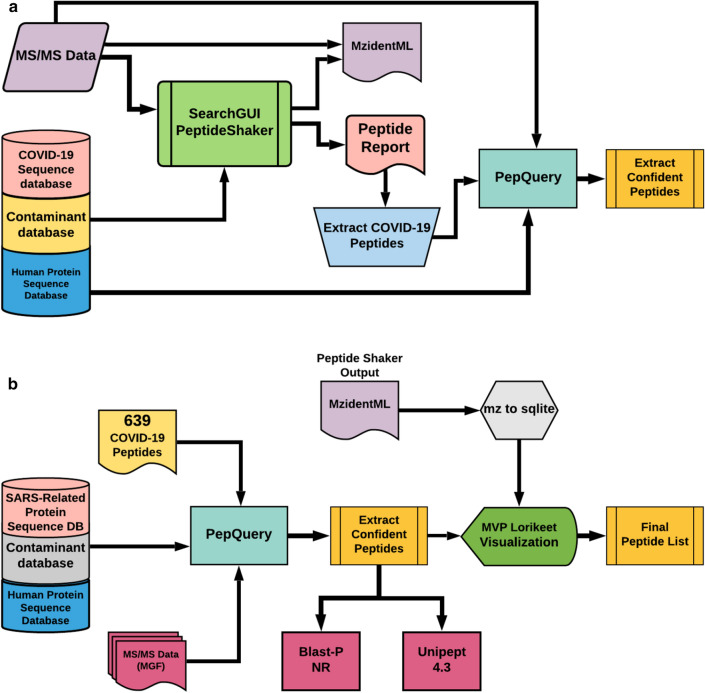
Fig. 2.
Workflows used in the interrogation of MS-data to identify and validate SARS-CoV-2 peptides (a) Galaxy-based sequence database searching workflow to detect and confirm SARS-CoV-2 peptides. MS/MS spectra from cell culture or clinical datasets were searched against appropriate protein sequence databases (protein sequences from COVID-19, contaminants, and Human Protein sequences) using SearchGUI/ Peptide Shaker. The peptide output was filtered to extract COVID-19 peptides and the output was confirmed using PepQuery to extract confident peptides. mzidentML generated through this workflow was subsequently used for analysis in Lorikeet (b) Workflow to validate detected SARS-CoV-2 peptides. A list of 639 Peptides (theoretical and validated peptides obtained from the cell-culture and clinical datasets) was subjected to PepQuery analysis of COVID-19 datasets to identify the presence of SARS-CoV-2 peptides. The quality of the peptide spectral matches (PSMs) was reviewed using Lorikeet visualization within the Multi-omics Visualization Platform for further validation. Peptides were also searched against NCBI-non redundant database and Unipept 4.3 for taxonomic annotation