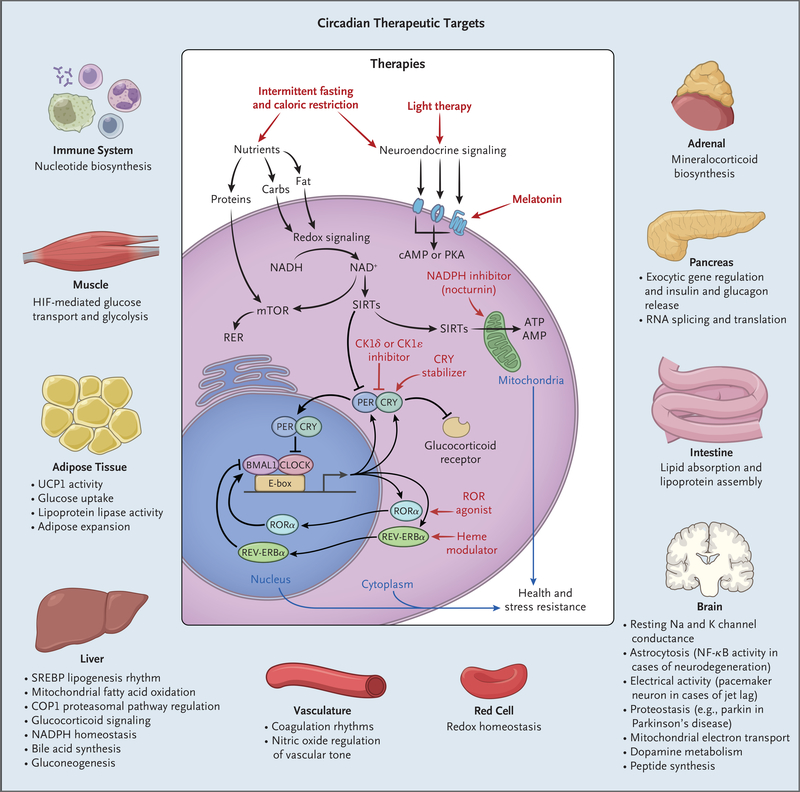
Figure 3. Potential Exploitation of Clock Pathways for Therapeutics.
Molecular clocks in nearly all cell types drive gene transcription in collaboration with tissue-specific factors. Local clocks generate robust oscillation in the expression of distinct rate-limiting factors according to the time of day. Therapies that affect either central clock activity (e.g., light or melatonin, each of which modulates the sleep–wake cycle) or peripheral-acting targets (e.g., modulators of nucleotide levels, cryptochrome [CRY] stability, and nuclear receptor activity) represent potential targets for manipulating circadian pathways in specific tissues. Shown are the pathways regulated in diverse tissues and, in the center, the localization of input signals and downstream therapeutic targets that are circadian outputs. Carbs denotes carbohydrates, cAMP cyclic AMP, CK1 casein kinase 1, ER endoplasmic reticulum, HIF hypoxia-inducible factor, mTOR mammalian target of rapamycin, NAD+ nicotinamide adenine dinucleotide, NF-κB nuclear f actor κB, PKA protein kinase A, SIRT sirtuin, and UCP1 uncoupling protein 1.