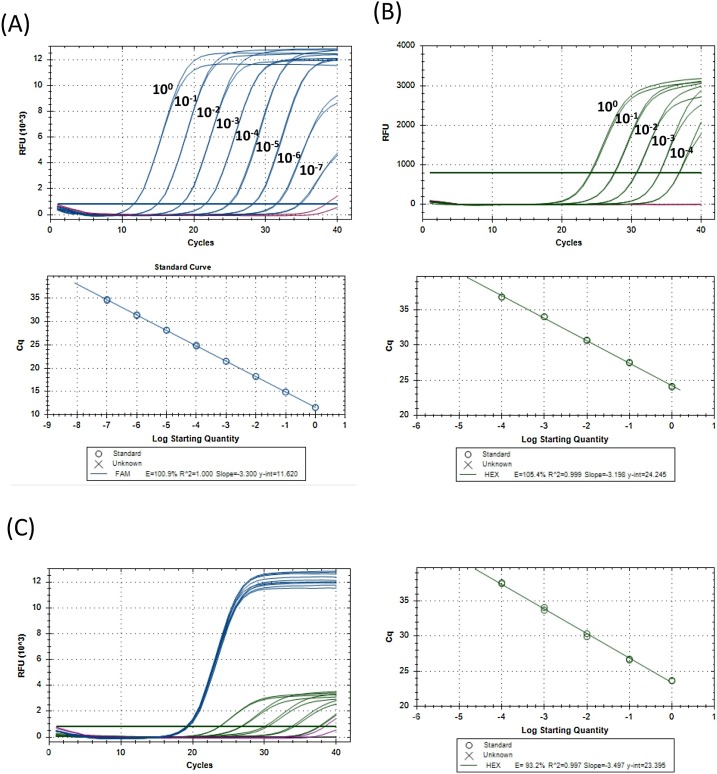
Fig. 1.
Linear range and PCR efficiency for E and RNase P.
RNA was extracted from SARS-CoV-2-infected Vero cells (A), serially diluted (10 fold) in nuclease-free water with carrier RNA and assayed in duplicate using the E/RNase P RT-qPCR assay. Relative fluorescence units (RFU) detected in the FAM channel for the E gene for samples containing viral RNA (blue) or carrier RNA alone (purple) are shown. (B) RNA was extracted from human peripheral blood mononuclear cells (PBMC), serially diluted (10 fold) in nuclease-free water with carrier RNA and assayed in duplicate using the E/RNase P RT-qPCR assay. RFU detected in the HEX channel for the RNase P for samples containing PBMC RNA (blue) or carrier RNA alone (purple) are shown. (C) RNA from SARS-CoV-2-infected Vero cells was spiked into serial dilutions of PBMC RNA. RFU from samples with PBMC and infected Vero cells RNA is shown in blue (FAM) and green (HEX), and carrier RNA alone is shown in purple (FAM) and black (HEX). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).