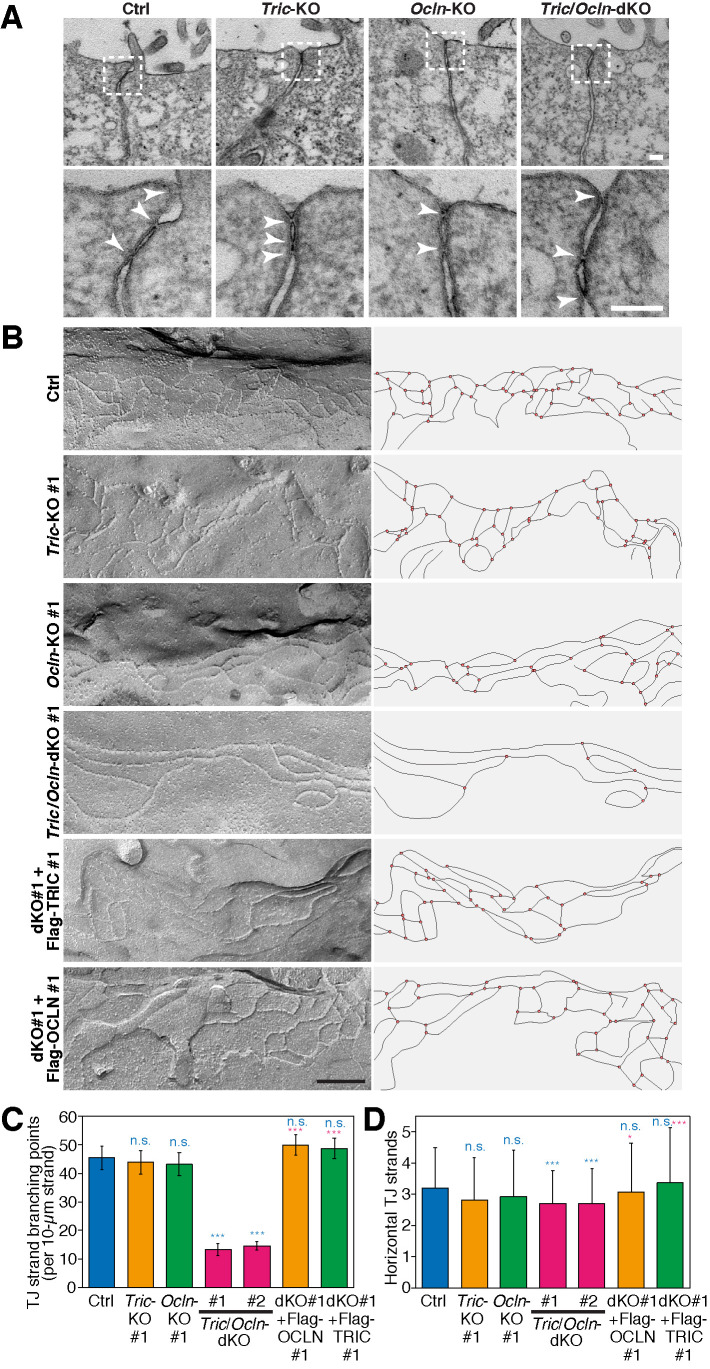
FIGURE 3:
The TJ-strand network of Tric/Ocln-dKO cells is less branched. (A) TEM images of Ctrl, Tric-KO #1, Ocln-KO #1, and Tric/Ocln-dKO #1 cells. The magnified images of the TJ regions in the top panels (dashed rectangles) are shown in the bottom panels. Kissing points between the adjacent cells are shown with white arrowheads in the bottom panels. Scale bars, 100 nm. (B) Freeze-fracture replica electron microscopy of Ctrl, Tric-KO #1, Ocln-KO #1, Tric/Ocln-dKO #1, dKO #1+Flag-OCLN #1, and dKO #1+Flag-TRIC #1 cells are shown (left panels). Additional images are shown in Supplemental Figure S6. Scale bars, 200 nm. TJ strands on the replica images were traced, and the network organization is shown (right panels). (C) Numbers of TJ-strand junction points (branching points). Number of three-way junction points were divided by total length of TJ strands (>100 µm from more than 20 images for each clone). Four-way junctions were regarded as two three-way junctions. Error bars indicate 95% confidence intervals. Total lengths of the measured TJ strands were 101 µm (Ctrl), 103 µm (Tric-KO #1), 103 µm (Ocln-KO #1), 127 µm (Tric/Ocln-dKO #1), 248 µm (Tric/Ocln-dKO #2), 155 µm (dKO #1+Flag-OCLN #1), 146 (dKO #1+Flag-TRIC #1). ***p < 0.001; n.s., p ≥ 0.05 in Poisson’s exact rate test. (D) Number of horizontal TJ strands. n = 188 (Ctrl), 184 (Tric-KO #1), 208 (Ocln-KO #1), 198 (Tric/Ocln-dKO #1), 244 (Tric/Ocln-dKO #2), 356 (dKO #1-Flag-OCLN #1), 310 (dKO #1-Flag-TRIC #1). ***p < 0.001; *p < 0.05; n.s., p ≥ 0.05 in Welch’s t test with Bonferroni’s correction. See Supplemental Figure S2, B and C, for detailed quantification methods for C and D.