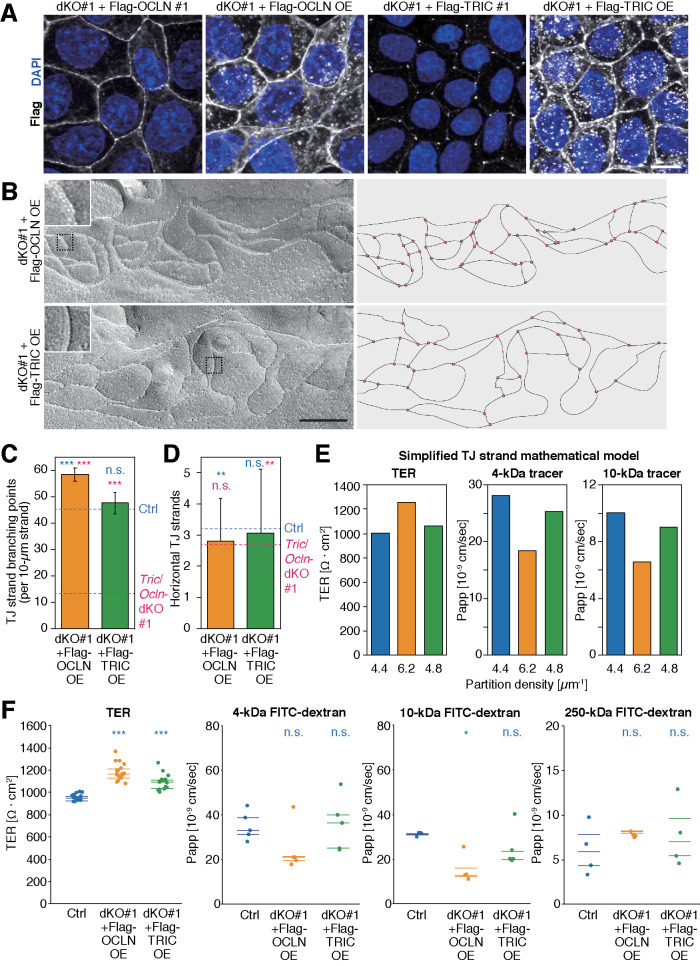
FIGURE 6:
Overexpression of occludin results in increased TJ network complexity and reduced permeability. (A) dKO #1+Flag-OCLN #1, dKO #1+Flag-OCLN OE, dKO #1+Flag-TRIC #1, and dKO #1+Flag-TRIC OE cells were stained with mouse anti-Flag mAb (white) and DAPI (blue). Scale bar, 20 µm. (B) Freeze-fracture replicas of dKO #1+Flag-OCLN OE and dKO #1+Flag-TRIC OE cells. Insets are magnified images of the region indicated with black dotted squares. Scale bar, 200 nm. (C) Numbers of TJ-strand junction points (branching points). Branching point numbers of Ctrl (cyan dashed line) and Tric/Ocln-dKO #1 (magenta dashed line) cells shown in Figure 3C are indicated for comparison. Error bars indicate 95% confidence intervals. Total lengths of the measured TJ strands were 100 µm (dKO #1+Flag-OCLN OE) and 284 µm (dKO #1+Flag-TRIC OE). ***p < 0.001; n.s., p ≥ 0.05 in Poisson’s exact rate test (vs. Ctrl cells [cyan] and vs. Tric/Ocln-dKO #1 cells [magenta]). (D) Number of horizontal TJ strands. Horizontal strand numbers of Ctrl (cyan dashed line) and Tric/Ocln-dKO #1 (magenta dashed line) cells shown in Figure 3D are indicated for comparison. n = 286 (dKO #1+Flag-OCLN OE) and 667 (dKO #1+Flag-TRIC OE). **p < 0.01; n.s., p ≥ 0.05 in Welch’s t test with Bonferroni’s correction (vs. Ctrl cells [cyan] and vs. Tric/Ocln-dKO #1 cells [magenta]). (E) Averaged TER values and Papp of 4- and 10-kDa macromolecule tracers in the simplified TJ-strand models with partition densities of 4.4 (Ctrl cell model), 6.2 (OCLN-OE cell model), and 4.8 µm–1(TRIC-OE cell model) and three horizontal rows. The Papp and TER values are an average of 512 and 106 simulations, respectively. (F) TER values and Papp of 4-, 10-, and 250-kDa FITC-dextran in the Ctrl, dKO #1+Flag-OCLN OE, and dKO #1+Flag-TRIC OE cells. n = 16–17 (TER), n = 4–5 (tracer flux); 25, 50, and 75 percentiles are shown (lines). ***p <0 .001; **p < 0.01; *p < 0.05; n.s., p ≥ 0.05 in Tukey–Kramer test.