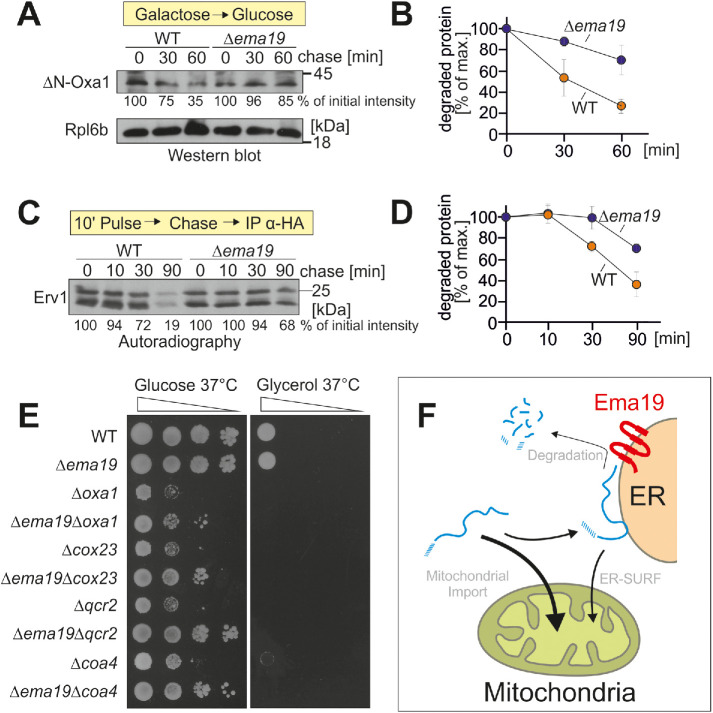
FIGURE 6:
Δema19 cells show reduced protein degradation. (A) The nonimportable ΔN-Oxa1 was expressed in WT and Δema19 cells. Expression was stopped by shifting cells from galactose to glucose for the times indicated (chase). Cells were isolated and the levels of ΔN-Oxa1 were analyzed by Western blotting and quantified. The ribosomal protein Rpl6b served as loading control. (B) The results from three biological replicates were quantified. Mean and standard deviations are shown. (C) Cells expressing HA-tagged Erv1 were grown to log phase. Radiolabeled methionine was added for 10 min (pulse). Cells were incubated in the presence of nonradioactive methionine (chase) for the times indicated. Cells were isolated, lysed, and used for immunoprecipitation with HA-specific antibodies (IP α-HA). The levels of the radiolabeled Erv1-HA protein were detected by autoradiography. (D) The results from three biological replicates were quantified. Mean and standard deviations are shown. (E) Cells of the indicated strains were grown on galactose and dropped onto glucose- or glycerol-containing plates. (F) Model for the role of Ema19 in the degradation of ER-associated mitochondrial precursor proteins.