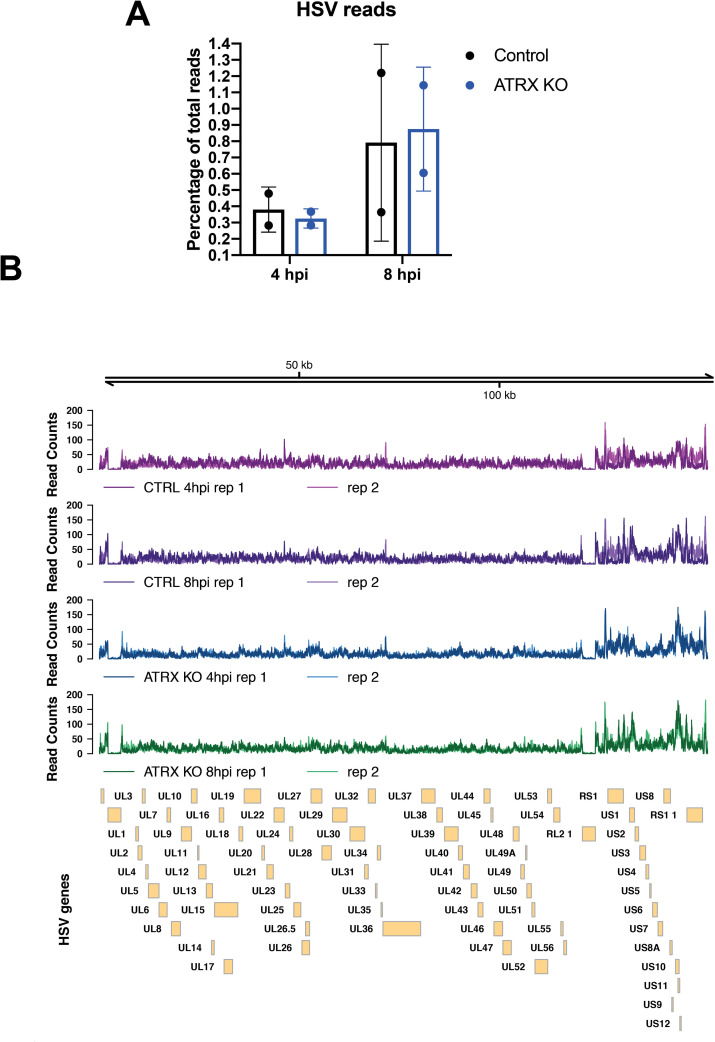
Fig 4. Levels of H3 on HSV genome in infected cells at 4 and 8 hpi by ChIP-seq. H3 ChIP-seq reads in ATRX-KO or Control cells infected with 7134 at MOI3 and harvested at 4 or 8 hpi were aligned to a human+HSV genome.
(A) The percentage of HSV reads to total aligned reads was calculated. (B) Alignments were visualized across HSV genome after normalizing reads to total HSV reads. Data shown in Fig 4 is from 2 independent experiments.