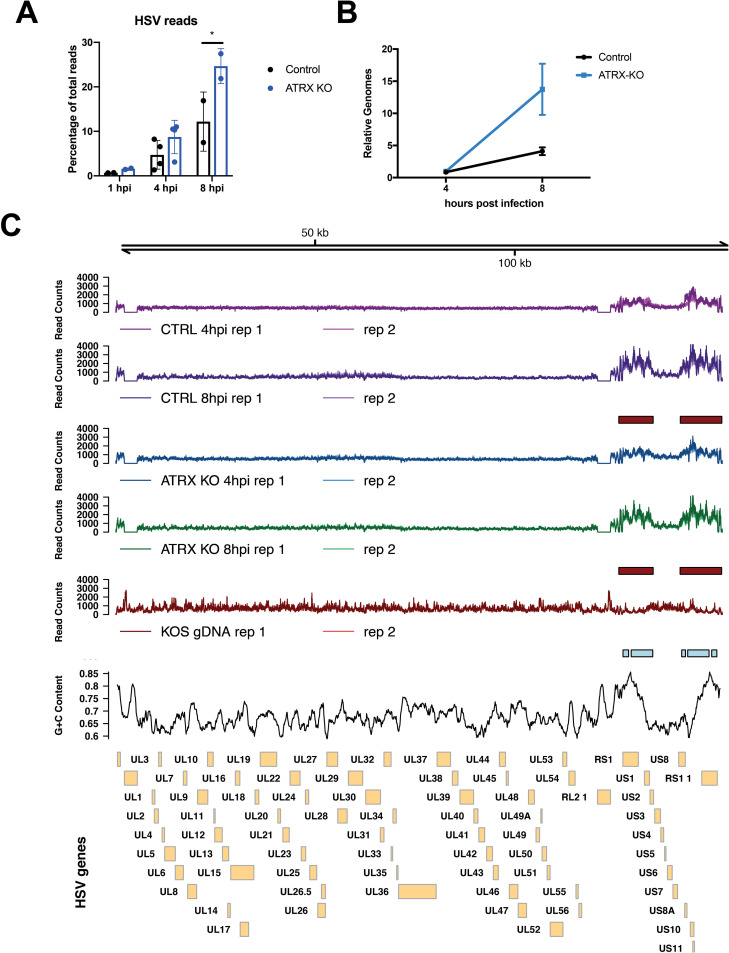
Fig 5. Accessibility of HSV DNA during infection as determined by ATAC-seq.
ATAC-seq was used to determine accessibility of the HSV genome in ATRX-KO or Control cells that were infected with an ICP0- HSV (7134) at MOI 5 and harvested at 1, 4, and 8 hpi. (A) Reads were aligned to a human+HSV genome and percentage of HSV reads to total mapped reads was calculated. (B) Relative HSV genomes were quantified by qPCR analysis of ICP8/GAPDH signals in TN5 treated ATRX-KO and Control samples at 4 and 8 hpi with 7134. Data are reported as the average of 2 independent experiments, except for the 4hpi data in (A) in which 4 replicates were included, ± standard error of the mean; p < 0.05 (*). (C) ATAC- seq reads that align to the HSV genome in samples from proteinase K treated HSV (KOS) genomes, Control cells and ATRX-KO cells infected with 7134 at MOI 5 and harvested at 4 and 8 hpi. Reads were normalized to total HSV reads prior to visualization. Boxes underneath tracks indicate regions of significant (FDR < 0.05) differential accessibility (red = increased accessibility, blue = decreased accessibility) between ATRX-KO cells infected with 7134 at 8 hpi vs 4 hpi, Control cells infected with 7134 at 8 hpi vs 4 hpi, and proteinase K-treated KOS genomes vs Control cells at 8 hpi. GC content in 1000 bp windows across HSV KOS genome is visualized for comparison.