Fig. 1 |. H1 depletion in the haematopoietic compartment results in reduced numbers of terminally differentiated lymphocytes in immune tissues.
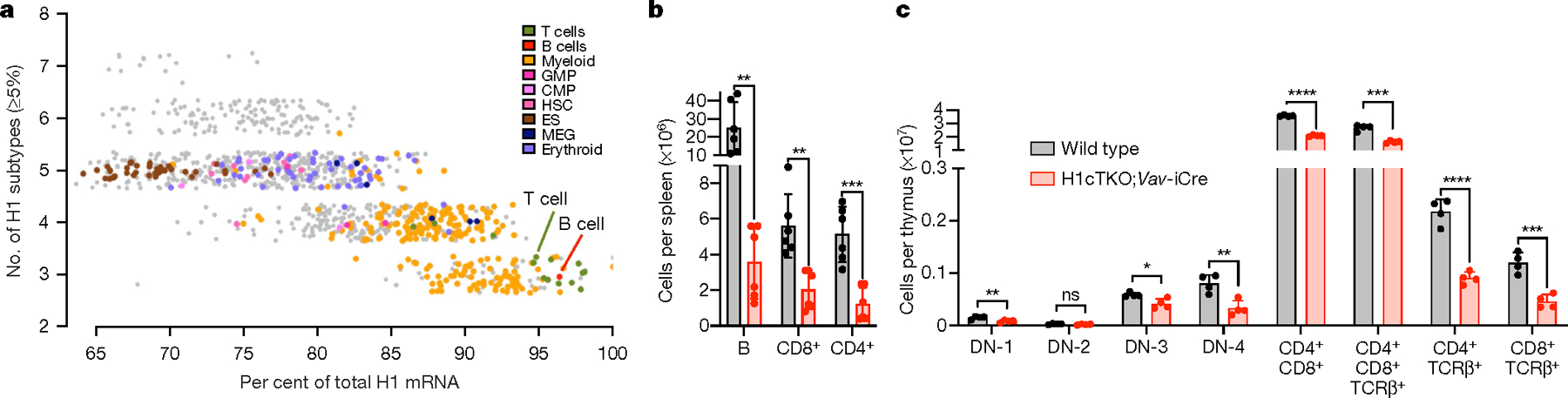
a, In silico analysis of H1 expression using mouse FANTOM56 cap analysis gene expression–sequencing (CAGE-seq) data. Total H1 was calculated as the sum of the relative log expression of normalized CAGE-seq reads mapping to the unique 5′ RNA sequences of the 11 H1 subtype transcripts. Shown is the percentage of total H1 corresponding to subtypes H1C, H1D and H1E as a function of the number of H1 subtypes expressed in each tissue. CMP, common myeloid progenitors; ES, embryonic stem cell; GMP, granulocyte–macrophage progenitors; HSC, haematopoietic stem cells; MEG, megakaryocytes. b, Number of B cells, CD4+ and CD8+ T cells in spleens (n = 6) of wild-type and H1cTKO;Vav-iCre mice, as measured by flow cytometry. c, The frequency of the indicated immature and mature T cell populations in the thymus of wild-type (n = 4) and H1cTKO;Vav-iCre (n = 4) mice was measured by flow cytometry. Absolute cell number was calculated by multiplying the relative frequency of each population by the mean thymus cellularity for wild-type and H1cTKO;Vav-iCre mice. Data are mean ± s.d.; unpaired t-test; *P ≤ 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
