Figure 2. The m6A reader YTHFD2 is required for maintenance of glioblastoma stem cells. See also Figure S2.
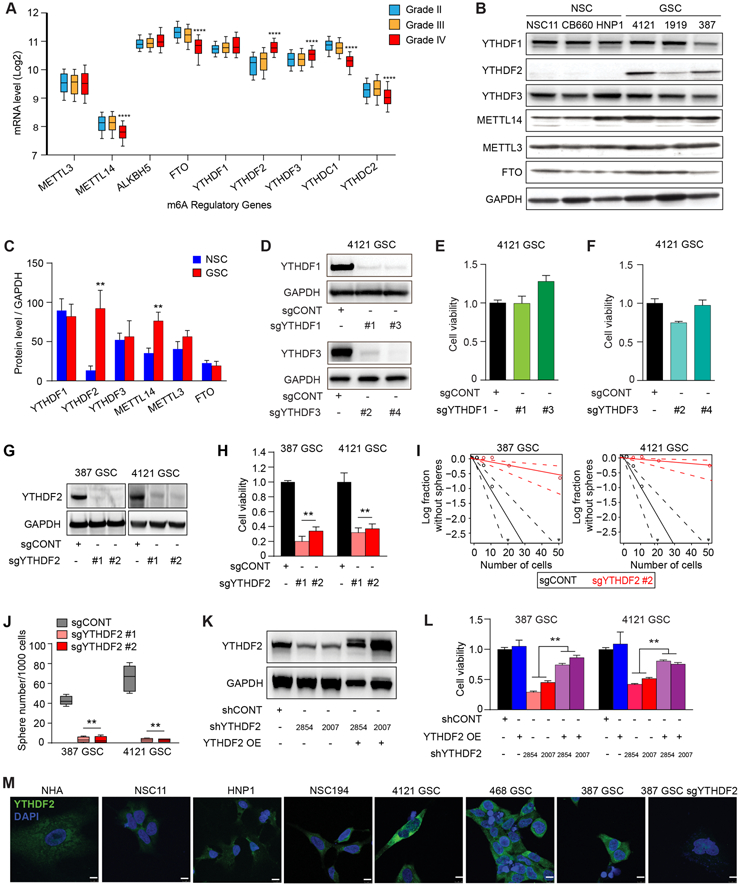
A, mRNA expression of selected m6A regulatory genes in the TCGA GBM-LGG dataset based on tumor grade. ****, p < 0.0001 for Grade IV vs either Grade II or III. B, Immunoblot assessment of YTHDF1, YTHDF2, YTHDF3, METTL14, METTL3, and FTO protein levels in NSCs and GSCs. GAPDH was used as a loading control. C, Densitometry graphs showing the protein levels of m6A readers, writers and erasers normalized to GAPDH in GSCs compared to NSCs. **, p < 0.01. Error bars show standard deviation. D, Immunoblot showing YTHDF1 and YTHDF3 levels after transduction of GSCs with a control non-targeting sgRNA sequence (sgCONT) or four independent sgRNAs targeting YTHDF1 (sgYTHDF1 #1, and sgYTHDF1 #3) and YTHDF3 (sgYTHDF3 #2 and sgYTHDF3 #4). E and F, Relative cell viability of 4121 GSCs expressing a control non-targeting sgRNA sequence (sgCONT) or two independent sgRNAs targeting YTHDF1 (sgYTHDF1 #1 and sgYTHDF1 #3) and YTHDF3 (sgYTHDF3 #2 and sgYTHDF3 #4). Error bars show standard deviation. G, Immunoblot showing YTHDF2 protein levels after transduction of two GSCs (387 and 4121), with a control non-targeting sgRNA sequence (sgCONT) or two independent sgRNAs targeting YTHDF2 (sgYTHDF2 #1 and sgYTHDF2 #2). H, Cell viability of two patient-derived GSCs (387 and 4121) following transduction with either a non-targeting control sgRNA (sgCONT) or one of two independent, non-overlapping sgRNAs targeting YTHDF2. **, p < 0.01. Error bars show standard deviation. I, Sphere formation using an extreme limiting dilution assay (ELDA) was performed with 387 and 4121 GSCs expressing sgCONT or sgYTHDF2 #2. J, Box plot shoeing the quantification of the number of spheres (per 1,000 cells) formed by GSCs. Data are presented as mean (+/−SEM) from three independent experiments. **, p < 0.01. K, Immunoblots demonstrating the knockdown and overexpression efficiency of YTHDF2 in 4121 GSCs. GAPDH is used as a loading control. L, Cell viability of GSCs transduced with one of two independent, non-overlapping shRNAs against YTHDF2 (shYTHDF2) or control shRNA (shCONT), with or without exogenous YTHDF2 overexpression. **, p < 0.01. Error bars show standard deviation. M, Cellular localization and levels of YTHDF2 in GSCs, NSCs and normal astrocytes as demonstrated by immunofluorescence staining using YTHDF2 antibody. Scale bars: 10 μm.
