Extended Data 1. Low-burden bacterial signal in fetal meconium.
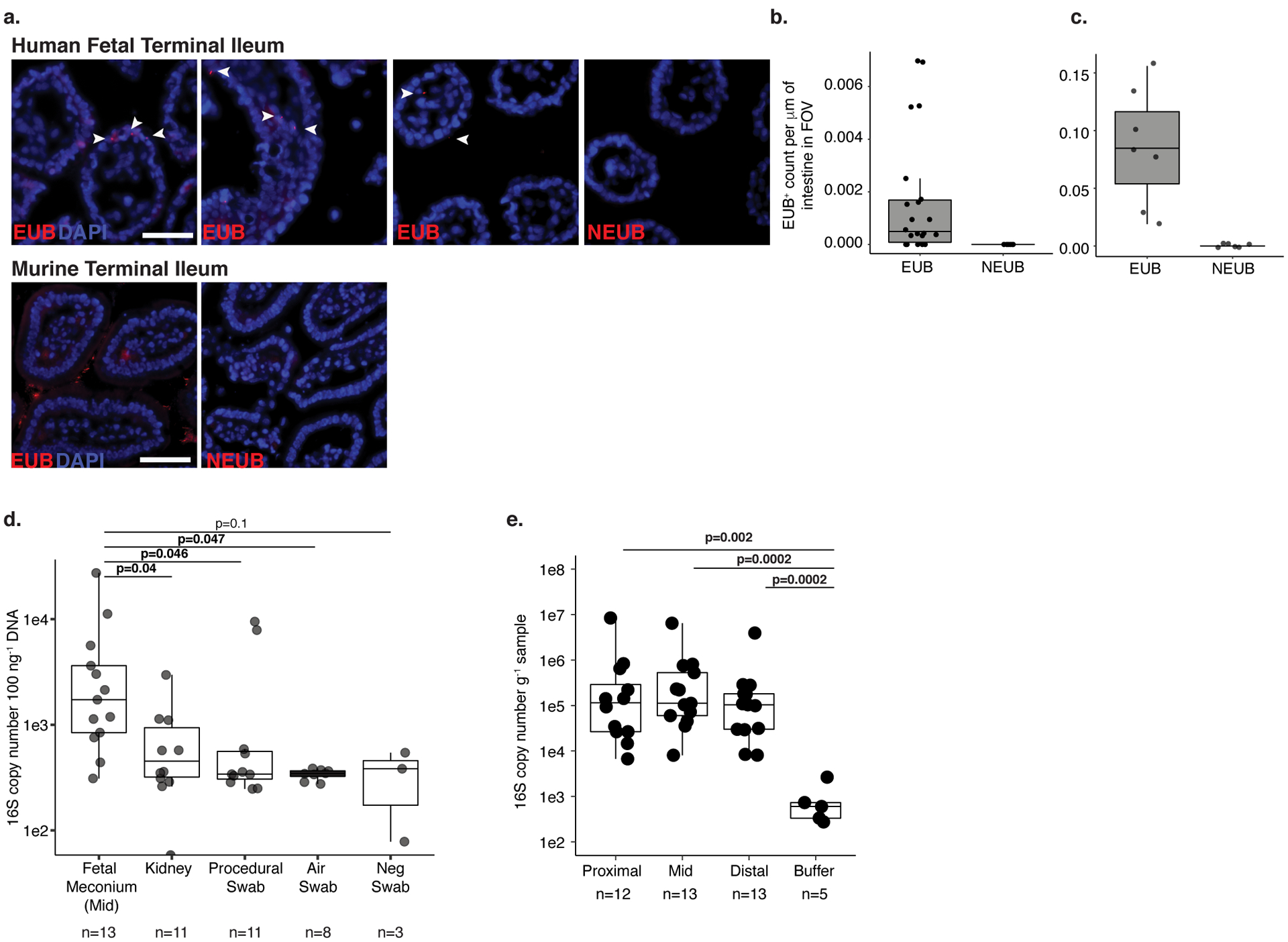
a. Fluorescent in situ hybridization probes targeting eubacteria (EUB) or non-targeting probe (NEUB) in 5 μm cryosections of human fetal (top panel) or murine (bottom panel) terminal ileum at 400x magnification. Arrowheads indicate EUB-positive findings in fetal sections. Scale bar corresponds to 50 mm. Quantification of independent fields of view (FOV) per mm within a representative b. human fetal (n=28) or c. murine (n=13) intestinal segments, where n is an independent segment of the intestine. For a-b, representative images of three fetal specimens and three mice. d. Total 16S copy number per 100 ng gDNA in meconium from mid-section of the fetal small intestine (n=13), fetal kidney (n=11), and procedural (n=11), air (n=8), or blank (n=3) swab was quantified by qPCR of DNA extracts using a standard curve where n represents biologically independent samples; two-sided Satterthwaite’s method on linear mixed effects model to test for significance. e. Total 16S copy number per gram frozen sample in meconium from proximal (n=12), mid (n=13), and distal (n=13) sections of the fetal small intestine where n represents independent biological samples across the length of the intestine or extraction buffer (n=5, n represents biologically independent samples) was quantified by qPCR of DNA extracts using a standard curve; two-sided Wilcoxon rank sum test for significance compared to buffer control. Boxplots indicate the median (center), the 25th and 75th percentiles, and the smallest and largest values within 1.5× the interquartile range (whiskers).
