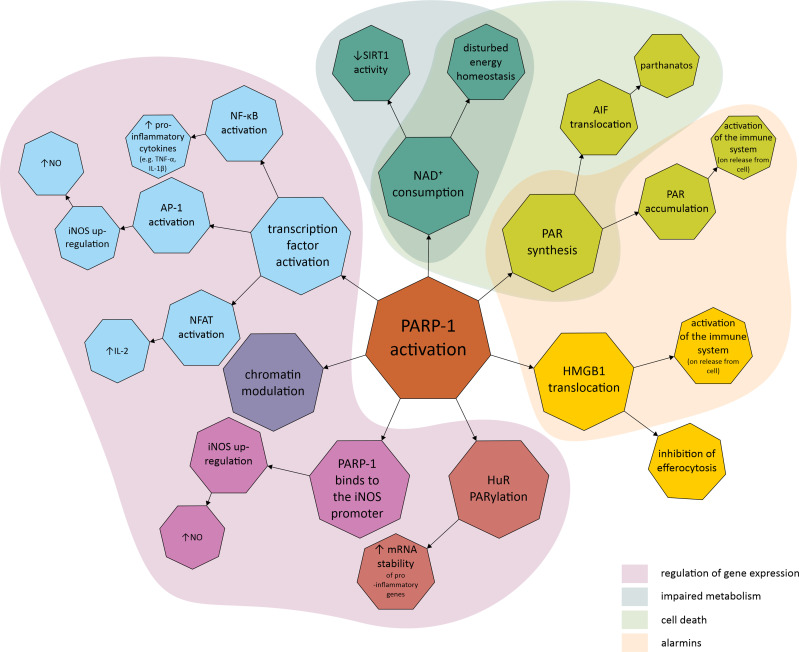
Figure 2.
Molecular mechanisms linking poly (ADP-ribose) polymerase (PARP) with inflammatory response and disturbed cell metabolism. The first area covers the mechanisms involved in the regulation of gene expression, such as PARP-1 promotion and activation of transcription factors, chromatin modulation, PARP-1 binding to the promoter region of the gene, and regulation of post-transcriptional modification as exemplified by HuR PARylation. The second area covers the mechanisms related to metabolic impairment, which include a decrease in SIRT1 activity and cellular energy depletion, both of which are related to NAD+ depletion due to over-activation of PARP-1. The third area contains mechanisms related to cell death, including parthanatos, PARP-1 dependent and caspase-independent cell death related to PAR accumulation and their interaction with AIF. The final area covers the mechanisms leading to the production of alarmins, endogenous molecules that activate the innate immune system when released from the cell due to, eg, cellular damage.
Abbreviations: AP-1, activator protein-1; HMGB1, high-mobility group box 1; HuR, human antigen R; IL, interleukin; iNOS, inducible nitric oxide synthase; mRNA, messenger ribonucleic acid; NAD+, nicotinamide adenine dinucleotide; NFAT, nuclear factor of activated T-cells; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NO, nitric oxide; PAR, poly (ADP-ribose); PARP-1, poly (ADP-ribose) polymerase 1; PARylation, poly ADP-ribosylation; SIRT1, sirtuin 1; TNF-α, tumour necrosis factor α.