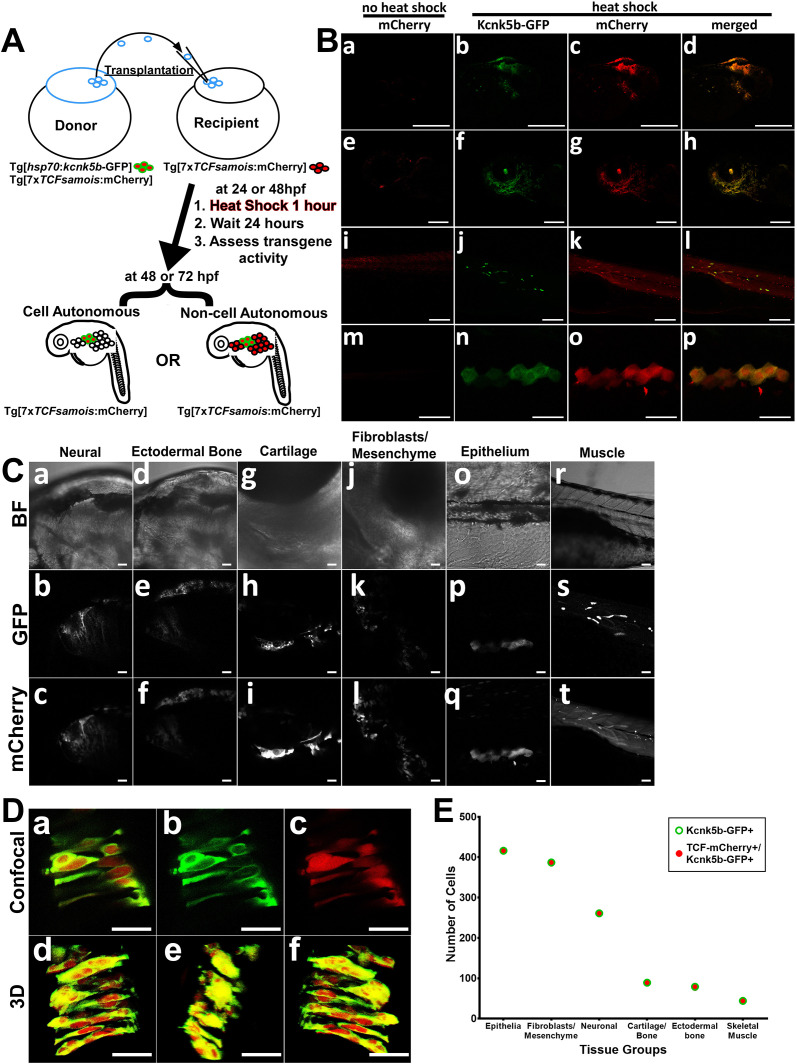
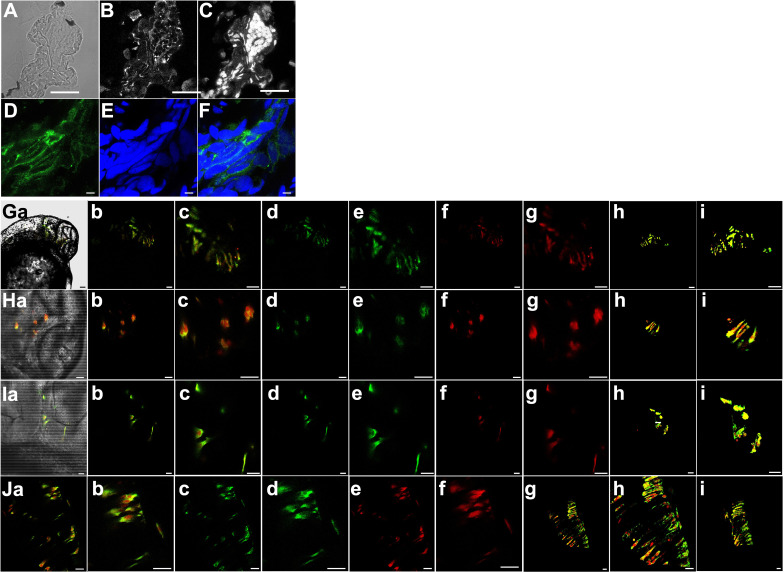
Figure 3. Kcnk5b induces Lef-1-dependent transcription in several tissues in a cell autonomous manner.
(A) Diagram of transplantation procedure and possible cell-autonomous and non-cell-autonomous outcomes on the expression of the Tg[7XTCF-Xla.sam:mCherry] Lef1 reporter after heat-shock induction of the Tg[hsp70:kcnk5b-GFP] transgene. (B) Transplantation experiments of donor cells from double transgenic fish harboring Tg[hsp70:kcnk5b-GFP] and Tg[7XTCF-Xla.sam:mCherry] into homozygous host embryos harboring only the Tg[7XTCF-Xla.sam:mCherry]. The head (a) eye (e) trunk (i) and finfold (m) of mosaic larva before heat shock induction of kcnk5b-GFP expression. Head (b–d), eye (f–h), trunk (j–l), and finfold (n–p) of 72 dpf larva at 24 hr after heat shock. (C) Bright field images of the head (a,d), jaw area (g) border tissue of otic vesicle (j) and trunk (o,r) of 72 hpf larva; GFP expression from Tg[hsp70:kcnk5b-GFP](b,e,h,k,p,s) and mCherry expression from Tg[7XTCF-Xla.sam:mCherry] (c,f,i,l,q,t). (D) Higher magnification of merged (a) and separate GFP (b) and mCherry (c) channels of cells in the neural tube of 48 hpf embryos. Z-stack composite three-dimensional images before rotation (d), rotated 90° (e) and 180° (f). (E) Total number of positive cells counted in the tissues of all mosaic larva for all mCherry-positive cells from recipient Tg[7XTCF-Xla.sam:mCherry] larva, all GFP-positive cells from Tg[hsp70:kcnk5b-GFP](open green triangle) and all double positive cells (open blue squares). The data for each experiment represent three or more experiments with two or more biological replicates. Scale bars are equal to 200 μm (Ba-l), 50 μm (Bm-p), 20 μm (C), and 25 μm (D).