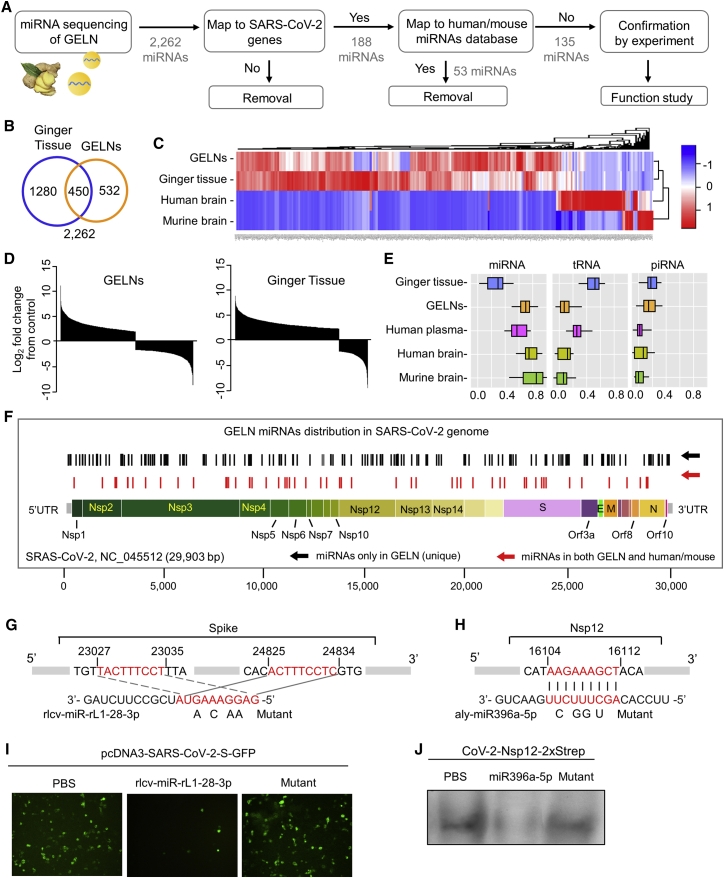
Figure 2.
GELN miRNA potentially targets to the RNA of the SARS-CoV-2
(A) Flow diagram depicting the steps taken in identifying unique GELN miRNAs potentially targeting the RNA of SARS-CoV-2. (B) Venn diagram of miRNAs detected in the ginger tissue and GELNs using miRNA sequencing. (C) Heatmap showing miRNAs from ginger tissue, GELNs, and human and mouse brain (n = 3 per group). (D) Waterfall plot showing the differences in the relative abundance of miRNAs between GELNs and ginger tissue normalized by human miRNAs. (E) Distribution of RNA biotype differences. Boxes represent median and interquartile ranges. (F) Schematic diagram and distribution of the putative binding sites of GELN miRNAs in the full-length SARS-CoV-2 genome. UTR, untranslated region. The miRNAs of humans and mice that have the same mapping seed sequences as GELNs are indicated in red and were excluded in further experiments. (G and H) Predicted consequential pairing of target region of spike gene (G, top), Nsp12 gene (H, top), GELN rlcv-miR-rL1-28-3p (G, bottom), and aly-miR396a-5p (H, bottom), respectively. The miRNA seed matches in the target RNAs are mutated at the positions as indicated. (I) A549 cells transfected with CoV-2 S inserted into pcDNA3-GFP and GELN rlcv-miR-rL1-28-3p, mutant RNA. Visualization with confocal fluorescence microscopy. (J) A549 cells transfected with Nsp12 inserted into pLVX-EF1alpha-2xStrep-IRES and GELN aly-miR396a-5p, mutant RNA. Visualization is with Strep-Tactin-HRP conjugate by immunoblot. Data are representative of three independent experiments (error bars, SD). ∗p < 0.05, ∗∗p < 0.01 (two-tailed t test).