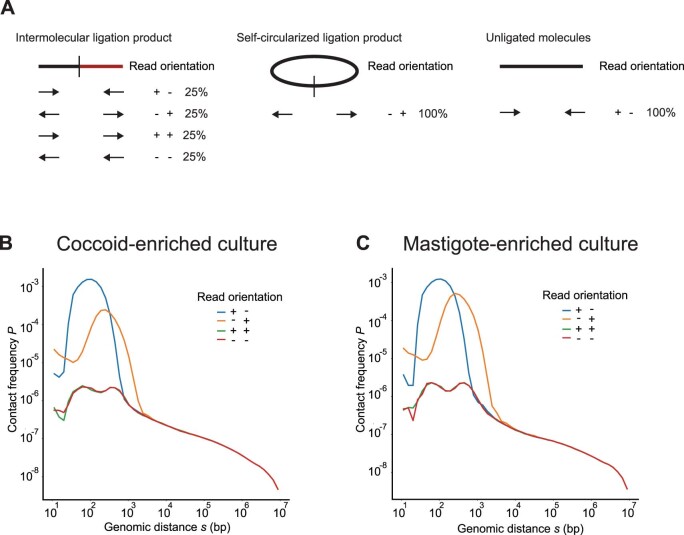
Extended Data Fig. 6. P(s) plots for different Hi-C read orientations.
a, Different types of Hi-C ligation products. Ligation of two interacting restriction fragments can occur in 4 different fragment orientation (head-to-head, tail-to-tail, head-to-tail and tail-to-head). These 4 products can be identified by the Hi-C read orientation, as indicated, and are expected to occur with equal frequency (25% for each type). Self-circularized (partial) digestion products, or unligated linear fragments are also produced in Hi-C and are uninformative. These two products are characterized by the two Hi-C read orientations for each molecule: (-) and (+) for self-circularized molecules and (+) and (-) for unligated molecules. b, Genome-wide P(s) plots for each of the 4 Hi-C read orientations for coccoid-enriched cultures. For distances below 2–3 kb uninformative Hi-C dominate, while for genomic distance over 2–3 kb all 4 read-orientation occur with equal frequency. Hi-C data was mapped against Smic1.0. c, Genome-wide P(s) plots for each of the 4 Hi-C read orientations for mastigote-enriched cultures. For distances below 2–3 kb uninformative Hi-C dominate, while for genomic distance over 2–3 kb all 4 read-orientations occur with equal frequency. Hi-C data was mapped against Smic1.0.