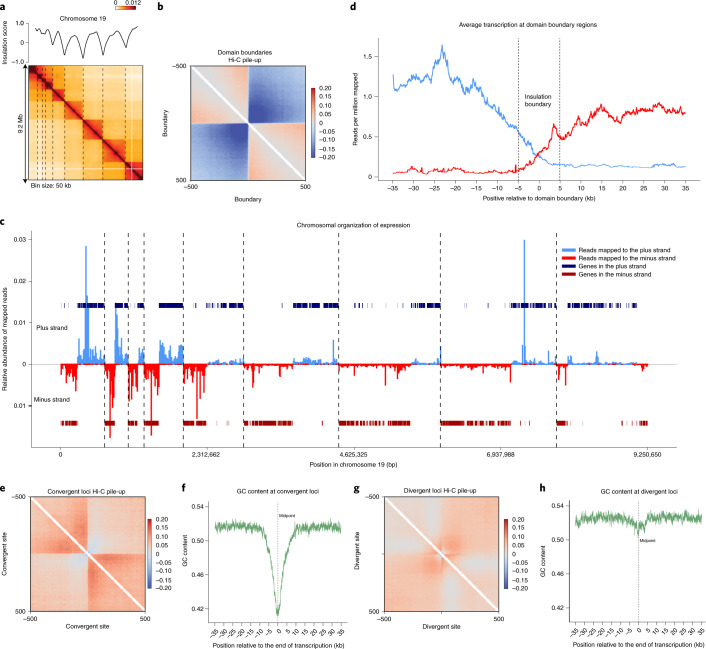
Fig. 5. Correlation between Hi-C domains and unidirectional gene blocks.
a, Hi-C interaction map for chromosome 19 (Smic1.0; bin size = 50 kb) for mastigotes. Dotted lines indicate domain boundaries. Plot on top of the heatmap represents the insulation profile (10-kb resolution, window size 500 kb). b, Average Hi-C interactions around the set of 441 domain boundaries (±500 kb) at 10-kb resolution. Data are log10(observed interaction frequency/expected interaction frequency). c, Chromosome 19 transcription and domain landscape. Indicated are: transcripts mapping to the plus strand (light blue), transcripts mapping to the minus strand (red), genes on the plus strand (dark blue), genes on the minus strand (dark red), and domain boundaries (dotted vertical lines). d, Average relative transcription around boundaries. Values are averaged across all domain boundaries in the genome for regions 30 kb upstream and downstream domain boundaries and in 100-bp sliding windows. Dotted lines delimit domain boundary, which is a 10-kb region based on Hi-C data. The limited amount of transcription observed to proceed through the boundary can be the result of the imprecision of boundary detection (10 kb). e, Average Hi-C interactions around 280 manually curated convergent sites (±500 kb), previously not identified as domain boundaries, at 10-kb resolution. Data are log10(observed interaction frequency/expected interaction frequency). f, GC content around all convergent sites. Values are averaged across all manually curated convergent sites in the genome for regions 35 kb upstream and downstream to the midpoint between two expression blocks (dotted line) in 100-bp sliding windows. Data are log10(observed interaction frequency/expected interaction frequency). g, Average Hi-C interactions around 517 manually curated divergent sites (±500 kb) at 10-kb resolution. Data are log10(observed interaction frequency/expected interaction frequency). h, GC content around divergent sites. Values are averaged across all manually curated divergent sites in the genome for regions 35 kb upstream and downstream to the midpoint between two expression blocks (dotted line) in 100-bp sliding windows.