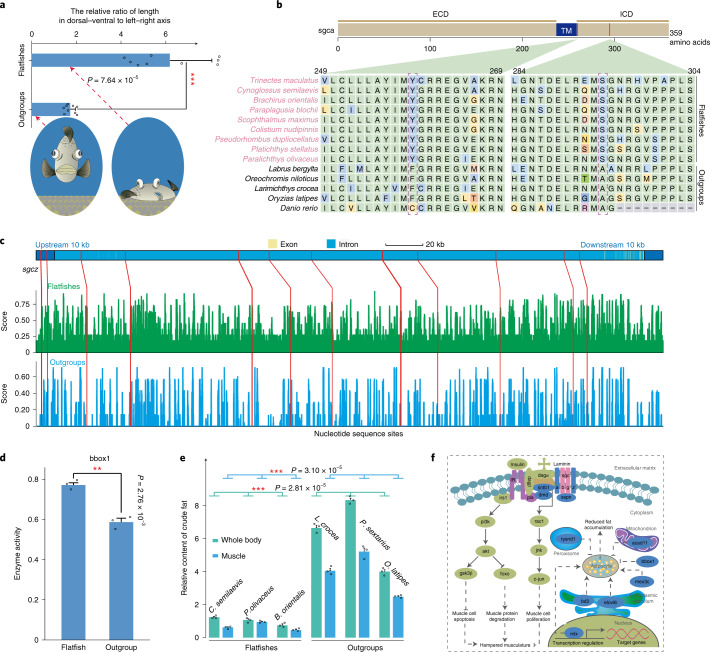
Fig. 3. Genetic changes correlated with the flat body plan in flatfishes.
a, The relative ratio of length in dorsal–ventral to left–right axis. The relative ratio of maximum height of dorsal–ventral axis/maximum length of left–right axis here was used to indicate the degree of body plan flatness of fishes. The ratio was measured in three individuals for each species, and the data are presented as mean values ±s.d. The statistical difference between groups was calculated using Student’s t-test (two tails) with *** representing a statistical P value <0.001. b, Mutations in RFP sgca compared to outgroups. ECD, ICD and TM represent the extracellular, intracellular and transmembrane domain, respectively. The fixed substitutions between RFP and outgroups are marked with a dashed box. c, SCNEs nearby sgcz gene. The x axis represents the nucleotide sequence sites across the sgcz gene and the y axis represents sequence similarity scores. The green and blue columns represent the average sequence similarity score within RFP and outgroups for each site, respectively; the red lines represent the physical locations of the SCNEs across the gene. d, The relative catalyzing activity of bbox1 in RFP and outgroups. The experiment was carried out three times and the data are presented as indicated above. e, Relative fat content in whole body and muscle tissues in flatfishes and outgroups. The fat content was measured in three individuals for each species, and the data are presented as indicated above. f, Hypothetical signaling pathway that may correlate with the body-plan flatness of flatfishes. Proteins marked blue are those encoded by genes that have undergone genetic alterations in RFP.