Fig. 6.
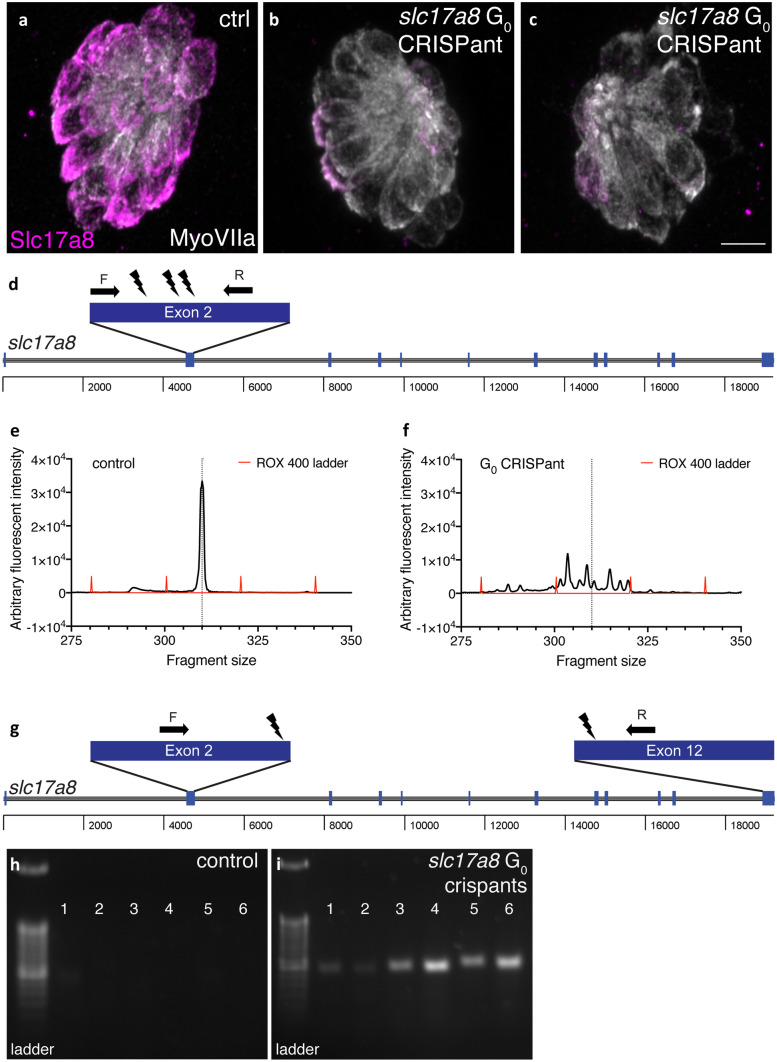
Two examples G0 slc17a8 CRISPant analysis and genotyping. Neuromasts from uninjected a and CRISPants embryos injected with the following gRNAs directed against the following sites in exon 2 of slc17a8 (5′-3′): GACAGAAGATGGTCGGCCGG (TGG), GGTGCTTTGGCCTTCCCAAA (CGG), and GCCCACCCCTATTGGACTGT (GGG) along with Cas9 protein b–c. Staining with anti-Slc17a8 (Obholzer et al. 2008) and anti-MyosinVIIA (Developmental Studies Hybridoma Bank, #138-1) to label lateral line hair cells reveal that Slc17a8 staining is absent in G0 CRISPants that lack an acoustic startle response. Schematic of PCR analysis of slc17a8 d used to detect INDELs. The CRISPR-STAT assay, relying on fluorescent fragment analysis can be used to genotype individual CRISPants larvae and test gRNA efficiency. In these examples, there is a single peak in control larvae at 310 bp e. By comparison, in G0 slc17a8 CRISPants the peak at 310 bp is degraded, and numerous fragments (indicative of the many INDELs present in this mosaic founder) surrounding this peak are present f. Schematic of PCR analysis of slc17a8 g used to detect a large deletion. This PCR analysis was conducted on genomic DNA from uninjected control and CRISPant larvae lacking a startle response. Primers flank the sites targeted by the guides targeting exon 2 ((5′-3′)CACAGTCTACATCAACGGGA(CGG)) and exon 12 (TCCAGTGTAATGCACCATGG(AGG)) and were used to amplify the region between exon 2 and exon 12. Deletion of a 14.2-kb region in CRISPants yielded an ~ 400-bp PR product (lanes 1–6, i) that was absent in uninjected controls (lanes 1–6, h). Images in a–c were taken at × 63 magnification on a Zeiss LSM 780 confocal microscope. Scale bar in c = 5 µm