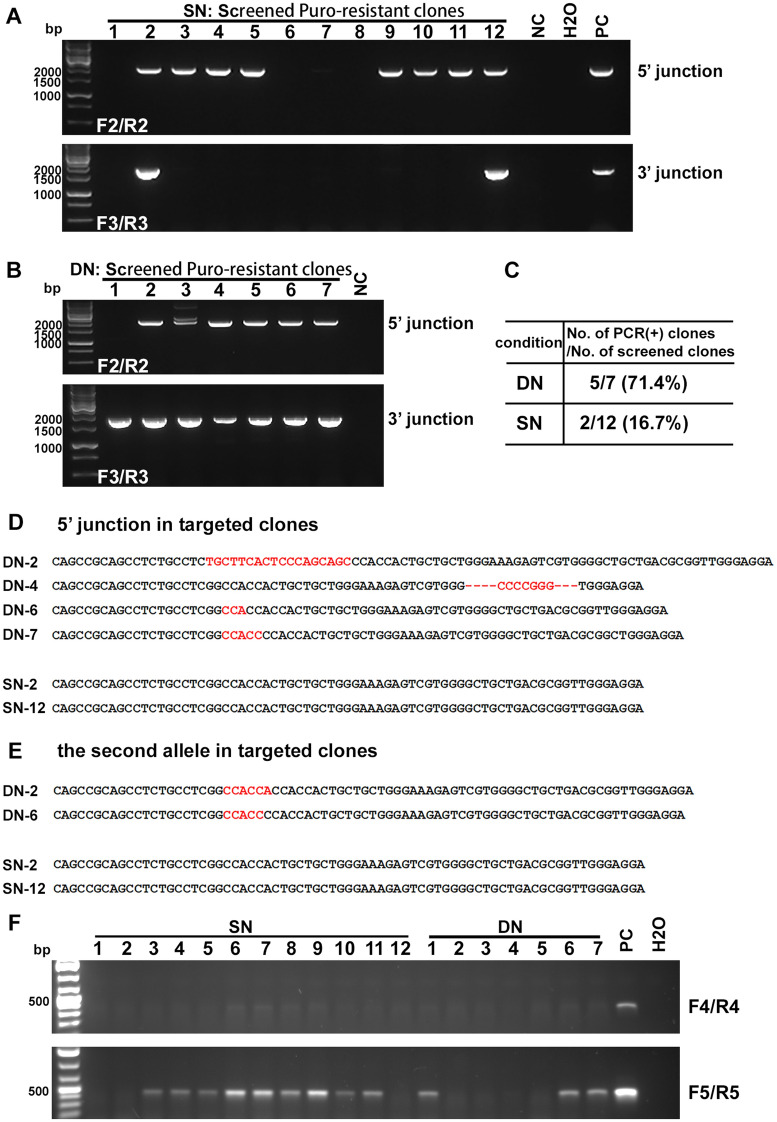
Figure 3.
Comparison of DN and SN efficiency and fidelity. (A,B) PCR-based screening of 5′ and 3′ junctions to identify successfully targeted iPSC clones for SN (A) and DN (B) treated cells. 1.9 kb band (primers F2/R2) indicates a successfully generated 5′ junction. 1.6 kb band (primers F3/R3) indicate a successfully generated 3′ junction. Primer locations are as indicated in Fig. 2A; unincorporated donor DNA and junctional regions of unsuccessfully edited cells cannot be amplified by these primers. PC: positive control; NC: negative control. (C) Summary of targeting efficiency. (D) DN, but not SN, is associated with indels in the edited allele. Sequence of the 5′ junction PCR product (primer F2/R2) was used for analysis. The corresponding sequence on the donor was used as the reference for comparison. Indels are in red. (E) DN, but not SN, is associated with indels in the second allele that does not undergo HDR. Sequence of PCR product (primer F1/R1) for the second allele was used for analysis. Sequence of the unedited parental line was used as the reference for comparison. Indels are in red. (F) Random donor integration in all screened SN and DN clones were examined by PCR using primers F4/R4 (upper image) and F5/R5 (lower image), which bind to the donor plasmid backbone. A diluted donor plasmid was used as the positive control (PC). More integration was detected with F5/R5 primers; SN-2 and -12 clones had no random integration. Primer locations as indicated in Fig. 2A.