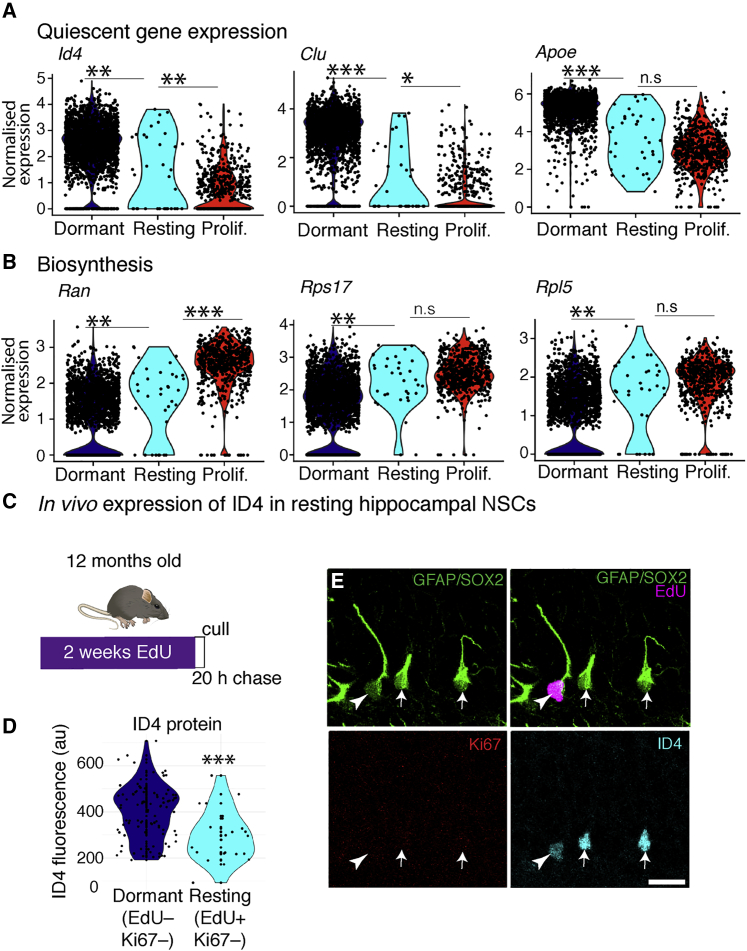
Figure 5.
Resting NSCs display a distinct transcriptional profile indicative of a shallow quiescent state
(A) Resting NSCs exhibit lower expression of quiescence marker genes than dormant NSCs (scRNA-seq).
(B) Resting NSCs have increased expression of genes involved in biosynthesis compared with dormant NSCs (scRNA-seq).
(C) To validate the differential gene expression analysis, 12-month-old mice received EdU for 2 weeks and were culled after a 20-h chase.
(D) Resting NSCs had reduced staining intensity for ID4 than dormant NSCs.
(E) Image of resting NSC (EdU+Ki67–, arrowheads) and dormant NSCs (EdU–Ki67–, arrows) with ID4 co-staining.
Graphs are violin plots. Dots in violin plots represent individual cells. Statistics: t test performed on Pearson’s residuals with false discovery rate (FDR)-corrected p value in (A) and (B) and t test in (D). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bar in (E), 15 μm. a.u., arbitrary units.