Fig. 2. The 3′UTR of STAT3 mRNA is associated with its localization to cell protrusions.
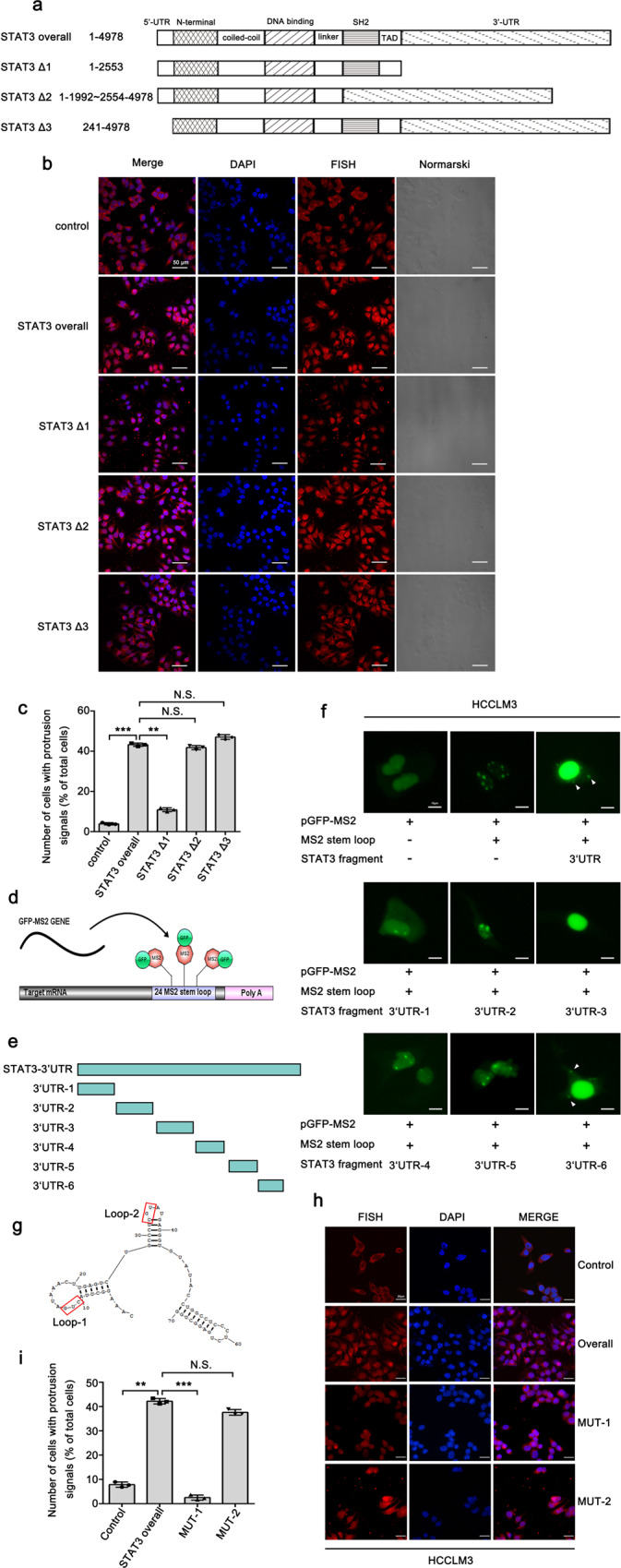
a Three fragments were constructed based on STAT3α full-length mRNA by deleting different functional domains. The length of fragments (1–4978) was numbered relative to the first nucleotide of the 5′-UTR. b FISH analysis images of STAT3 mRNA expression in control and STAT3 fragments-deleting cells, indicating that overexpression of STAT3 overall, STAT3 ∆2 and STAT3 ∆3 fragment could promote the protrusion-localization of STAT3 mRNA. Scale bar: 50 μm. c Quantitative analysis of b. Number of cells with protrusion signals reported as a percentage of total cells. d Schematic diagram showing the principle of the GFP-MS2 system set-up for detecting cytoplasmic localization element. e Six short fragments (3′UTR-1, 3′UTR-2, 3′UTR-3, 3′UTR-4, 3′UTR-5, 3′UTR-6) were constructed from STAT3 3′UTR and separately inserted into the GFP-MS2 system. f Immunofluorescence analysis of the GFP-MS2 system inserted with six short fragments from STAT3 3′UTR. STAT3 3′UTR and STAT3 3′UTR-6 can induce the cytoplasm GFP signal in the cell protrusions (indicated by arrow). Scale bar: 10 μm. g The secondary structure of STAT3 3′UTR-6 analyzed by RNA structure software showing the potential association with the localization of STAT3 mRNA. h Nucleotide substitution mutants were constructed in two different sites of STAT3 3′UTR-6 (mutation of loop1 termed as MUT-1, mutation of loop2 termed as MUT-2). The effect of site mutations on the protrusion-localization of STAT3 mRNA was evaluated by FISH assay. Scale bar: 25 μm. i Quantitative analysis of h. Number of cells with protrusion signals reported as a percentage of total cells. The values in the graphs represent the mean of three biologically independent experiments. Error bars represent ±s.d. *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed Student’s t-test.
