Abstract
The reduction of insulin resistance or improvement of insulin sensitivity is the most effective treatment for type 2 diabetes (T2D). We previously reported that Nogo-B receptor (NGBR), encoded by the NUS1 gene, is required for attenuating hepatic lipogenesis by blocking nuclear translocation of liver X receptor alpha, suggesting its important role in regulating hepatic lipid metabolism. Herein, we demonstrate that NGBR expression was decreased in the liver of obesity-associated T2D patients and db/db mice. NGBR knockout in mouse hepatocytes resulted in increased blood glucose, insulin resistance, and beta-cell loss. High-fat diet (HFD)/streptozotocin (STZ)-treated mice presented the T2D phenotype by showing increased nonesterified fatty acid (NEFA) and triglyceride (TG) in the liver and plasma and increased insulin resistance and beta-cell loss. AAV-mediated NGBR overexpression in the liver reduced NEFA and TG in the liver and circulation and improved liver functions. Consequently, HFD/STZ-treated mice with hepatic NGBR overexpression had increased insulin sensitivity and reduced beta-cell loss. Mechanistically, NGBR overexpression restored insulin signaling of AMPKα1-dependent phosphorylation of AKT and GSK3β. NGBR overexpression also reduced expression of endoplasmic reticulum stress-associated genes in the liver and skeletal muscle to improve insulin sensitivity. Together, our results reveal that NGBR is required to ameliorate T2D in mice, providing new insight into the role of hepatic NGBR in insulin sensitivity and T2D treatment.
Keywords: NGBR, insulin, diabetes, beta-cell, ER stress, AMPK, triglyceride, animal model, liver
Abbreviations: AAV, adeno-associated virus; AMPKα, adenosine monophosphate activated protein kinase alpha; Atf4/6, activating transcription factor 4/6; BIP, binding immunoglobulin protein or 78 kDa glucose-regulated protein (GRP78); Glcn, D-(+)-glucosamine hydrochloride; GSK3β, glycogen synthase kinase 3 beta; HFD, high-fat diet; IRE1α, inositol-requiring enzyme 1 alpha; LXRα, liver X receptor alpha; NGBR, Nogo-B receptor; PERK, pancreatic endoplasmic reticulum kinase; Skip, skeletal muscle and kidney-enriched inositol polyphosphate phosphatase; STZ, streptozotocin
Diabetes mellitus is a chronic, progressive disease characterized by elevated blood glucose levels. Blood glucose control, the objective of diabetes treatment, is important to reduce the progression of diabetic complications. The majority of patients with diabetes are affected by type 2 diabetes (T2D), which results from the body’s ineffective use of insulin (1). Therefore, it is crucial for T2D to reduce insulin resistance or improve insulin sensitivity. The liver plays a central role in coordinating the whole-body metabolism. The increasing evidence suggests that hepatic insulin resistance is sufficient to induce several components of insulin resistance syndromes (2).
Nonalcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases. A strong association between lipid accumulation and T2D has been demonstrated (3). More than 70% of T2D patients have NAFLD while almost all nonalcoholic steatohepatitis patients exhibit insulin resistance independently of bodyweight (4), because the excessive hepatic lipid in turn enhances insulin resistance and evidence is growing that beta-cell function is impaired through lipotoxicity (5).
The reticulon-4 (RTN4), also known as neurite outgrowth inhibitor or Nogo, includes three isoforms (Nogo-A, B, C) by alternative promoter usage and splicing. NGBR was initially identified as a putative receptor for binding Nogo-B in endothelial cells, and binding of Nogo-B to NGBR can lead to endothelial cell proliferation/vascular remodeling and angiogenesis during development (6). NGBR is a transmembrane receptor that contains a conserved hydrophobic pocket, which regulates membrane localization of the prenylated protein. It also promotes the membrane accumulation of Ras by directly binding the prenylated Ras at the plasma membrane, resulting in promoting growth factors-stimulated tyrosine kinase pathways (7). Our previous study demonstrated that NGBR expression is decreased in human fatty liver tissue (8). Consistently, NGBR deficiency in mouse liver results in remarkable lipid accumulation, indicating that loss of NGBR may be a potential trigger for inducing hepatic steatosis. Indeed, our study further demonstrated that NGBR deficiency impairs the activation of adenosine monophosphate-activated protein kinase alpha (AMPKα) and promotes the nuclear translocation of liver X receptor alpha (LXRα), a nuclear receptor of activating fatty acid synthesis. In contrast, activation of AMPKα by metformin, the first-line medicine used for T2D treatment, blocks NGBR deficiency-induced LXRα activation and hepatic lipid accumulation, suggesting that NGBR-AMPKα pathway is required for blocking LXRα nuclear translocation and LXRα-mediated lipogenesis (8).
As shown in our previous study (8), NGBR hepatic-specific knockout mice have substantial increase of nonesterified fatty acid (NEFA) levels in the liver and plasma, which may affect insulin sensitivity. Herein, we further elucidated the pathophysiological role of NGBR in regulating insulin resistance in the context of T2D. To mimic the process of T2D in humans, we chose a mouse T2D model by high-fat diet (HFD) feeding and repeated injection of streptozotocin (STZ) at a low dose, which has been used in many studies (9, 10). In the early stage of T2D, the mass of beta cells expands, increasing insulin production to compensate for insulin insensitivity, which can be aggravated by HFD (11). Along with the development of T2D, the increased fatty acids in the beta cells can activate FOXO1, resulting in enhanced apoptosis of the beta cells (12). STZ is a naturally occurring alkylating antineoplastic agent that is particularly toxic to the insulin-producing beta cells of the pancreas in mammals. However, such kind of the beta-cell loss caused by STZ in mice is severer than humans and consistent with what happens to patients with the advanced stage of T2D (13, 14). Thus, repeated STZ injection plus HFD feeding has been suggested to be used for establishing the T2D animal model (9, 10).
In this study, we used this T2D mouse model to determine the effects of NGBR overexpression on insulin resistance and demonstrated that liver NGBR could be required for maintaining insulin sensitivity and preventing T2D development.
Results
Reduced NGBR expression in the liver is associated with insulin resistance
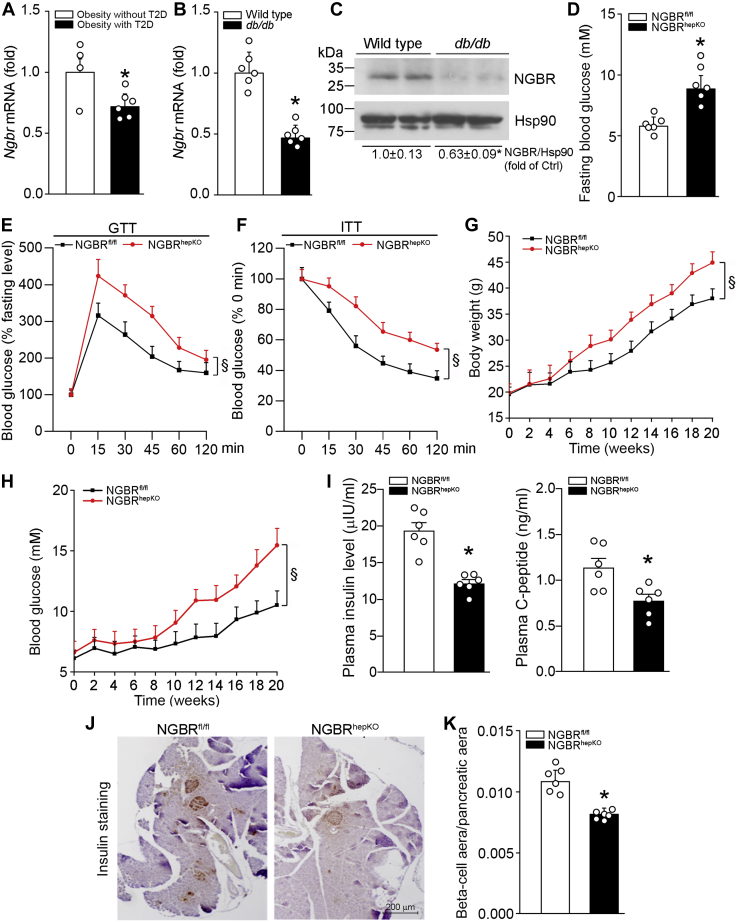
We searched the NCBI gene expression omnibus database (GEO access: GSE15653) and found that NGBR transcript level in the liver is decreased in obese-T2D patients as compared with non-T2D subjects (Fig. 1A). We further used quantitative real-time RT-PCR (qPCR) and western blot to determine NGBR transcript and protein levels in db/db mouse liver. Similarly, NGBR expression in db/db mouse liver is significantly decreased compared with normal controls (Fig. 1, B and C). These preliminary data suggest that NGBR may play an important role in T2D development. To test this hypothesis, we first examined the plasma glucose level in NGBR floxed littermate control (NGBRfl/fl) and NGBR hepatocyte-specific knockout (NGBRhepKO) mice under fasting conditions. As shown in Figure 1D, loss of NGBR expression in hepatocytes resulted in ∼40% increase in blood glucose level. Furthermore, glucose tolerance test (GTT) and insulin tolerance test (ITT) results revealed that severe insulin resistance occurred to NGBRhepKO mice (Fig. 1, E and F), the animals have hepatic steatosis due to activated LXRα-mediated fatty acid synthesis (8). With HFD feeding, NGBRhepKO mice showed increased bodyweight (Fig. 1G) and blood glucose levels (∼15.5 mM, Fig. 1H). The mice also showed decreased plasma insulin and C-peptide levels (Fig. 1I). Correspondingly, the decreased insulin-positive beta-cell mass was determined in the pancreases of NGBRhepKO mice (Fig. 1, J and K). Interestingly, NGBRhepKO mice also showed increased tumor necrosis factor α (TNFα) and interleukin 1β (IL1β) mRNA levels in the liver (Fig. S1). These data strongly suggest that reduced NGBR expression in the liver may be one of the major contributors to the etiology of T2D by reducing insulin production and inducing chronic inflammation.
Figure 1.
Reduced NGBR expression in the liver is associated with insulin resistance and loss of pancreatic beta-cells.A, NGBR transcript level is decreased in T2D obese patients. The data is generated from NCBI gene expression omnibus database (GEO access: GSE15653) ∗p < 0.05. n = 4 (Obesity without T2D group), n = 6 (Obesity with T2D group). B and C, expression of NGBR mRNA and protein in the liver of wild-type and db/db mice was determined by qPCR and western blot analysis. ∗p < 0.05, n = 6. D, NGBRfl/fl and NGBRhepKO mice at 12-week-old were randomly divided into two groups (n = 6). Blood glucose levels were determined after 12 h fasting. ∗p < 0.05, n = 6. E, glucose tolerance test (GTT): blood glucose levels were determined at the indicated time points after i.p. injection of glucose (0.5 g/kg bodyweight). §p < 0.05, n = 6. F, insulin tolerance test (ITT): the mice were preadministrated glucose (0.5 g/kg bodyweight) for 2 h after 12-h fasting, then blood glucose levels were determined at the indicated time points after i.p. injection of insulin (1 U/kg bodyweight). §p < 0.05, n = 6. G and H, NGBRfl/fl and NGBRhepKO mice at 8-week-old were fed high-fat diet (HFD) for 20 weeks. Bodyweight and blood glucose were determined at the indicated time points. §p < 0.05, n = 6. I–K, 8-week-old mice were fed HFD for 4 weeks. Plasma was collected and levels of insulin and C-peptide were determined by ELISA kits (I). The mass of beta cells in the pancreas was determined by immunohistochemistry staining of the tissue sections with anti-insulin antibody, followed by quantitative analysis of the ratio of beta-cell area to pancreatic tissue area (J and K). Scale bar: 200 μm (J). ∗p < 0.05, n = 6.
Overexpression of hepatic NGBR attenuates HFD-induced hepatic lipid accumulation
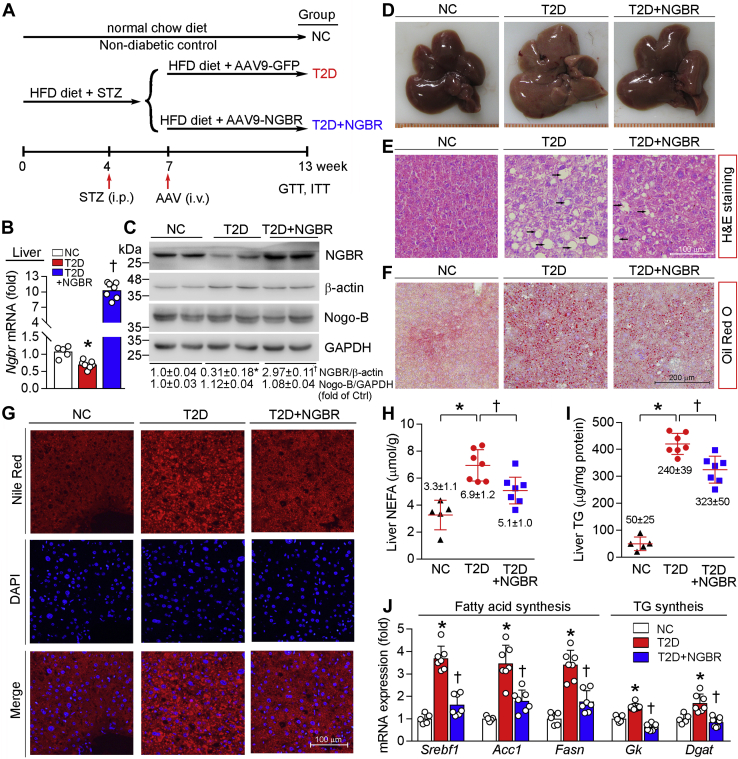
Next, we used HFD/STZ mouse diabetes model to determine the effect of NGBR overexpression on insulin resistance. Mice were fed HFD for 4 weeks and then i.p. injected STZ for seven consecutive days. Seven days after the first STZ injection, blood glucose reached 15 mM. Two weeks later, mice with high blood glucose were further divided into two groups and i.v. injected empty adeno-associated virus (AAV9) vector (AAV9-GFP, this group was named as T2D group) or AAV9-carrying NGBR gene (AAV9-NGBR, this group was named as T2D+NGBR group), and continued HFD feeding for another 6 weeks (Fig. 2A). At the end of study, we found that NGBR expression in T2D mouse liver was moderately decreased (Fig. 2, B and C), which is consistent with the results in db/db mice (Fig. 1, B and C). It also implies the link between liver NGBR expression and T2D.
Figure 2.
Overexpression of hepatic NGBR attenuates high-fat diet (HFD)/streptozotocin (STZ)-induced hepatic lipid accumulation.A, experimental design: male C57BL/6 mice at 5-week-old were randomly divided in two groups and fed normal chow or HFD. Four weeks later, HFD-fed mice were fasted for 6 h and then i.p. injected STZ (50 mg day−1 kg−1 bodyweight) daily for seven consecutive days. Normal chow-fed mice were injected same volume of vehicle (citrate buffer) and used as negative control (NC). After confirming onset of diabetes (3 weeks after the first STZ injection), STZ-injected mice were further randomly divided into two groups, followed by i.v. injection of adeno-associated virus 9 (AAV9)-GFP or AAV9-NGBR once at a dose of 1 × 1012 vg/mouse, and named as T2D group and T2D+NGBR group, respectively. B–J, at the end of experiment, mouse liver samples were used to conduct the following assays. Hepatic Ngbr mRNA was determined by qPCR (B). NGBR and Nogo-B protein expression was determined by western blot (C). Liver photograph (D). H&E, Oil red O, and Nile red staining of liver frozen sections (E–G). Black arrows indicate the vacuoles of lipid droplets (E). Scale bar: 100 μm (E and G), 200 μm (F). Hepatic nonesterified fatty acid (NEFA) and triglyceride (TG) levels were determined with total lipid extract from a piece of liver using corresponding assay kits and normalized to liver protein content (H and I). mRNA expression profiles in NC, T2D, and T2D+NGBR mouse liver including genes related to fatty acid synthesis: sterol regulatory element binding transcription factor 1 (Srebf1), acetyl-CoA carboxylase 1 (Acc1) and fatty acid synthase (Fasn), and TG synthesis: glycerol kinase (Gk) and diacylglycerol acyltransferase (Dgat), were determined by qPCR (J). ∗p < 0.05 versus NC; †p < 0.05 versus T2D. n = 5 (NC group), n = 7 (T2D or T2D+NGBR group).
Although Nogo-B has been considered as a putative ligand for NGBR binding, we determined that NGBR overexpression had little effect on Nogo-B expression in the liver (Fig. 2C). Reciprocally, NGBR deficiency had no effect on hepatic Nogo-B protein level and secretion in vitro and in vivo either (Fig. S2), indicating they might be independent molecules and have no interaction except in endothelial cells (6, 15). In T2D group, the palpable color of liver (Fig. 2D) and the presence of vacuoles in H&E staining sections (Fig. 2E) were determined, indicating the HFD feeding results in severe lipid accumulation. The Oil Red O staining and Nile Red staining further confirm the increased lipid accumulation and lipid droplets formed in the liver of T2D group (Fig. 2, F and G). The levels of NEFA and triglyceride (TG) were significantly increased in the liver of T2D mice as compared with nondiabetic mice [negative control (NC) group: wild-type mice were fed standard chow] (left halves of Fig. 2, H and I).
Compared with T2D group, infection of AAV9-NGBR substantially increased NGBR expression in the liver (Fig. 2, B and C). However, the levels of Ngbr mRNA in other tissues such as epididymal white adipose tissue (eWAT), skeletal muscle, and pancreas were not changed (Fig. S3), confirming the selective NGBR overexpression in the liver by AAV9-mediated gene delivery. Compared with NC group, STZ injection decreased mouse bodyweight but had little effect on food intake (Fig. S4). In addition, both bodyweight and food intake were similar between mice in T2D+NGBR and T2D groups during the treatment. Interestingly, high expressing NGBR reduced accumulation of NEFA and TG in the liver (Fig. 2, H and I), which were associated with reduced serum TG and NEFA levels (Table 1). The color of the liver of mice receiving AAV9-NGBR infection (T2D+NGBR group) was similar to that appeared in negative control group (NC) (Fig. 2D). The liver in T2D+NGBR group has reduced number of vacuoles with much smaller size in H&E staining, less lipid accumulation in Oil Red O staining, and fewer lipid droplets formed in Nile Red staining as compared with the liver in T2D group (Fig. 2, E–G). As shown in Table 1, HFD feeding increased levels of alkaline phosphatase (ALP), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and total bilirubin (TBIL), which are indicators of liver functional injuries. AAV9-mediated NGBR overexpression also improved liver functions by decreasing levels of AST, ALT, and TBIL. Mechanistically, AAV9-NGBR attenuated the HFD-induced expression of the genes involved in fatty acid synthesis, such as sterol regulatory element binding transcription factor 1 (Srebf1), acetyl-CoA carboxylase 1 (Acc1), and fatty acid synthase (Fasn); and the genes for TG synthesis, such as glycerol kinase (Gk) and diacylglycerol acyltransferase (Dgat) (Fig. 2J). These data further demonstrate that NGBR expression in the liver is essential for attenuating hepatic steatosis.
Table 1.
High expressing NGBR ameliorates diabetes-induced metabolic disorders
| Parameter | Group |
||
|---|---|---|---|
| NC | T2D | T2D+NGBR | |
| Triglyceride (TG) (mM) | 1.43 ± 0.32 | 3.24 ± 0.8a | 2.23 ± 0.46b |
| Nonesterified fatty acid (NEFA) (μM) | 341.14 ± 83.12 | 863.56 ± 430.5a | 598.78 ± 150.89b |
| Alkaline phosphatase (ALP) (U/l) | 57.97 ± 7.86 | 86.5 ± 15.21 | 82.68 ± 5.48 |
| Aspartate aminotransferase (AST) (U/l) | 175.91 ± 23.3 | 343.89 ± 74.58a | 231.63 ± 13.2b |
| Alanine aminotransferase (ALT) (U/l) | 36.17 ± 4.26 | 207.98 ± 65.23a | 123.46 ± 15.26b |
| Total bilirubin (TBIL) (μM) | 1.13 ± 0.2 | 2.14 ± 0.46a | 1.28 ± 0.51b |
| Blood urea nitrogen (BUN) (mM) | 8.06 ± 0.48 | 7.27 ± 0.58 | 6.54 ± 0.49 |
At the end of treatment as indicated in Figure 2A, mouse serum samples were prepared and used to complete biochemical analysis. Data are expressed as mean ± SD (n = 5, NC group; n = 7 for other two groups).
p < 0.05 versus NC.
p < 0.05 versus T2D.
Overexpression of NGBR in the liver improves insulin sensitivity
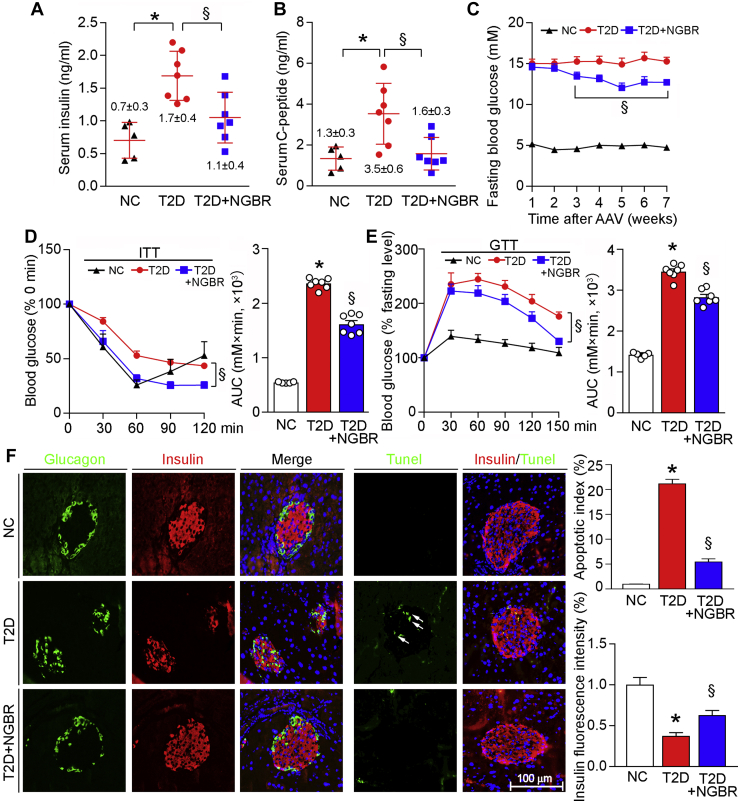
Meanwhile, we determined the effects of NGBR overexpression on glucose homeostasis and insulin sensitivity in the liver. T2D is associated with high fasting plasma glucose and insulin levels. Indeed, compared with nondiabetic mice (NC group), levels of fasting serum insulin, C-peptide, and glucose were significantly increased in T2D group (Fig. 3, A–C). In contrast, NGBR overexpression in the liver substantially reduced them (Fig. 3, A–C). To further assess the effects of NGBR overexpression on whole-body glucose homeostasis and insulin resistance, we performed ITT and GTT. Compared with T2D group, we observed decreases in blood glucose levels 30 min after glucose injection (for ITT) and 30 to 60 min after insulin injection (for GTT), as well as AUC for glucose in T2D+NGBR group, which may reflect an increase in insulin sensitivity in mouse liver by NGBR overexpression (Fig. 3, D and E). Taken together, the results in Figure 3, A–E suggest that AAV-mediated NGBR overexpression in the liver ameliorates HFD/STZ-induced glucose intolerance and insulin resistance.
Figure 3.
Overexpression of hepatic NGBR improves insulin sensitivity.A and B, at the end of experiment as indicated in Figure 2A, mouse blood samples were collected and determined serum insulin and C-peptide levels by ELISA. C–E, the experimental design is shown in Figure 2A. After viral injection, fasting blood glucose was determined weekly (C), and insulin tolerance test (ITT) (D) or glucose tolerance test (GTT) (E) was performed 9 or 5 days before the end of experiment as follows: the 4-h fasted mice were i.p. injected insulin (1 U/kg bodyweight) for ITT, while the 12-h fasted mice were oral administrated glucose solution (0.5 g/kg bodyweight) for GTT. Blood glucose levels were determined at the indicated time points after insulin or glucose administration with quantitation of area under curve (AUC). F, paraffin sections prepared from pancreas samples of mice used in Figure 2A were conducted immunofluorescent staining with anti-glucagon (green) or insulin (red) antibody, and TUNEL staining respectively. White arrows indicate apoptotic cells. Scale bar: 100 μm. ∗p < 0.05 versus NC; §p < 0.05 versus T2D. n = 5 (NC group), n = 7 (T2D or T2D+NGBR group).
As shown in Figure 1, J and K, lack of NGBR expression in the liver reduced insulin-positive beta-cell mass in mouse pancreas. Accordingly, we examined whether hepatic NGBR overexpression can protect islet against damage or beta-cell loss occurring to the diabetic animal model. Compared with nondiabetic mice, the results of glucagon and insulin immunofluorescent staining in Figure 3F demonstrated reduced islet size and increased pancreatic alpha cells in pancreases of diabetic mice, indicating severe damage to islets occurs. However, NGBR overexpression prevented reduction of islets size, loss of beta cells, and infiltration of alpha cells (Fig. 3F). In addition, the number of apoptotic beta cells determined by TUNEL staining was decreased in T2D+NGBR group (Fig. 3F), indicating that hepatic NGBR overexpression can also protect against beta-cell apoptosis in T2D. As a negative regulator of hepatic fatty acid synthesis (8), NGBR overexpression in the liver may contribute to improved beta cells by reducing circulating NEFA levels.
NGBR regulates hepatocyte insulin sensitivity through insulin signaling, AMPK axis
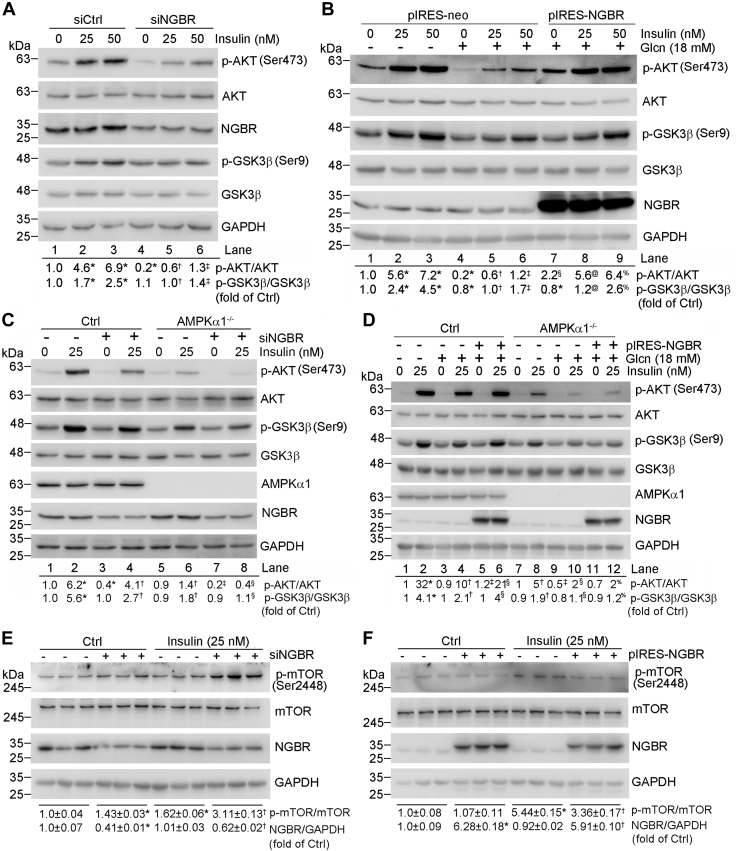
Insulin activates insulin receptors mainly by activating AKT/PKB and PKCζ cascades (16, 17). Activated AKT induces glycogen synthesis through inhibition of GSK3β by phosphorylating it. To unveil the mechanisms by which NGBR enhances insulin sensitivity, we knocked down NGBR expression in HepG2 cells and primary hepatocytes by transfecting cells with NGBR siRNA and then treated cells with insulin. We found that reduced NGBR expression led to decreased phosphorylated AKT (p-AKT) at the basal level in HepG2 cells (Fig. 4A, lane 4 versus 1), and primary hepatocytes isolated from wild-type mice (Fig. S5). Furthermore, we found the insulin-stimulated phosphorylation of AKT and GSK3β (p-GSK3β) were also decreased in siNGBR-transfected cells (Fig. 4A, lane 2/3 versus 5/6, respectively), indicating that NGBR should be involved in regulating insulin sensitivity. To investigate if NGBR also affects insulin sensitivity in the insulin-resistant state, we treated cells with glucosamine hydrochloride (Glcn), a precursor of the hexosamine biosynthetic pathway (HBP), which can mimic the high-glucose-induced insulin resistance at the cellular level (18, 19, 20). After Glcn treatment, phosphorylation of AKT was decreased (Fig. 4B, lane 4 versus 1), which was reversed by NGBR overexpression (Fig. 4B, lane 7 versus 4 and 1). In addition, the effects of insulin on induction of phosphorylation of AKT or GSK3β were impaired by Glcn (Fig. 4B, lane 5 or 6 versus 2 or 3), and the Glcn-caused impairment was blocked by NGBR overexpression (Fig. 4B, lane 8 or 9 versus 5 or 6).
Figure 4.
NGBR regulates insulin sensitivity through insulin signaling, adenosine monophosphate-activated protein kinase (AMPK) axis, and inhibiting mTOR in HepG2 cells.A and C, HepG2 cells (A) or HepG2-Ctrl cells and HepG2-AMPKα1−/− cells (C) in 6-well plates were transfected with scrambled siRNA (siCtrl, 50 nM) or NGBR siRNA (siNGBR, 50 nM) for 24 h. Cells were then treated with insulin at the indicated concentrations for 30 min. B and D, HepG2 cells (B) or HepG2-Ctrl cells and AMPKα1−/− cells (D) in 6-well plates were transfected with NGBR expression vector (pIRES-NGBR, 1 μg/well) or empty vector (pIRES-neo, 1 μg/well) for 12 h. Cells were then pretreated with glucosamine hydrochloride (Glcn) (18 mM) for 18 h, followed by insulin treatment for 30 min at the indicated concentrations. E, HepG2 cells in 6-well plates were transfected with scrambled siRNA (siCtrl, 50 nM) or NGBR siRNA (siNGBR, 50 nM) for 24 h. Cells were then treated with insulin (25 nM) for 30 min. F, HepG2 cells in 6-well plates were transfected with NGBR expression vector (pIRES-NGBR, 1 μg/well) or empty vector (pIRES-neo, 1 μg/well) for 12 h. Cells were then treated with insulin (25 nM) for 30 min. Expression of indicated proteins was determined by western blot. ∗p < 0.05 versus lane 1, siCtrl or pIRES-neo; †p < 0.05 versus lane 2, siNGBR or pIRES-NGBR; ‡p < 0.05 versus lane 3; §p < 0.05 versus lane 4; @p < 0.05 versus lane 5; %p < 0.05 versus lane 6. n = 3.
AMPK is an important sensor for cellular energy balance by regulating a wide array of physiological events including hyperglycemia and insulin sensitivity (21). To further assess the role of AMPK in NGBR-improved insulin sensitivity, we established an AMPKα1 knockout (AMPKα1−/−) HepG2 cell line and determined that AMPKα1 knockout decreased phosphorylation of AMPK target genes, such as ACC and Raptor (Fig. S6A). We also found that insulin-stimulated phosphorylation of AKT or GSK3β was decreased in AMPKα1−/− HepG2 cells. Inhibition of NGBR expression by siRNA further reduced both p-AKT and p-GSK3β (Fig. 4C), as well as p-AMPKα1 and p-IRS1 (Fig. S6B).
Next, we investigated the effect of NGBR overexpression on Glcn-impaired insulin signaling in both control and AMPKα1−/− HepG2 cells. In the insulin-treated HepG2 control cells, NGBR overexpression restored Glcn-impaired p-AKT (left halves, Fig. 4D). In contrast, NGBR overexpression cannot restore Glcn-impaired p-AKT in insulin-treated AMPKα1−/− HepG2 cells. These results suggest that NGBR regulates insulin signaling via AMPKα1.
mTOR (mechanistic target of rapamycin) is a key mediator of the insulin signaling pathway (22). We found that reduced NGBR expression led to increase of mTOR phosphorylation in HepG2 cells with or without insulin treatment (Fig. 4E). Reciprocally, NGBR overexpression reduced mTOR phosphorylation (Fig. 4F). These results suggest that reduced mTOR phosphorylation may contribute to NGBR-enhanced insulin signaling.
NGBR sensitizes insulin actions by improving endoplasmic reticulum (ER) stress
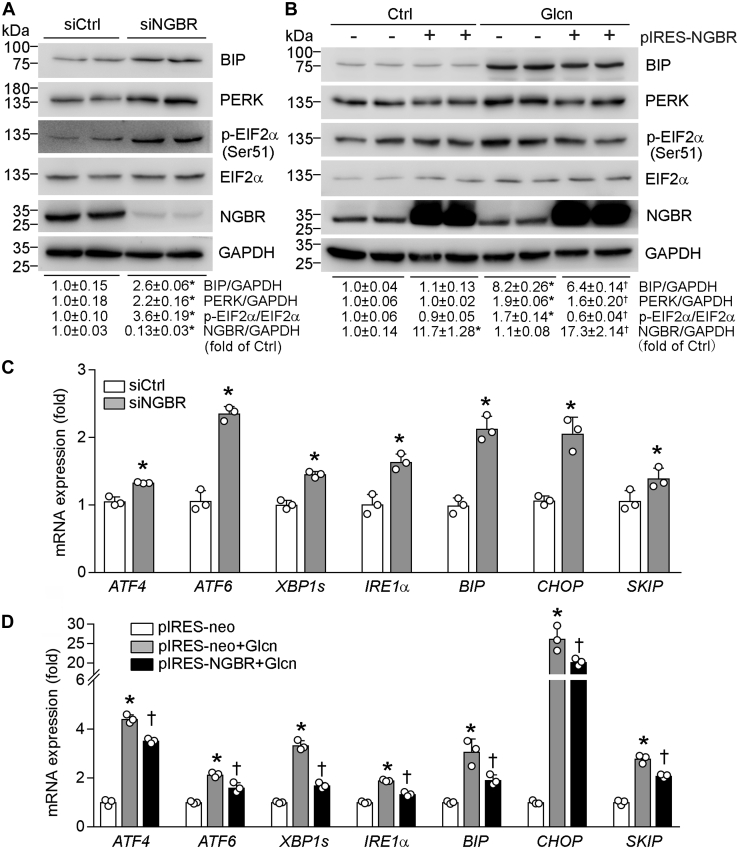
Glcn- or high-glucose-induced ER stress contributes to insulin resistance (23, 24). To investigate the role of NGBR in regulating ER stress, we first knocked down NGBR expression in HepG2 cells by NGBR siRNA and determined the effect of reduced NGBR on expression of ER stress-related genes. NGBR knockdown substantially increased mRNA and protein levels of binding immunoglobulin protein (BIP) (Fig. 5, A and C) as well as mRNA of DNA-damage inducible transcript 3 (CHOP) (Fig. 5C), indicating that reduction of NGBR expression may cause unfolded protein response (UPR). UPR is initiated by three ER transmembrane proteins: inositol-requiring enzyme 1α (IRE1α), pancreatic endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) (25, 26). We found that PERK protein (Fig. 5A) and mRNA of IRE1α and ATF6 were increased in NGBR knockdown HepG2 cells (Fig. 5C). The IRE1α RNase can excise a 26-nt intron from X-box binding protein 1 (XBP1) mRNA, which is then translated into XBP1s protein (27, 28). XBP1 translocates into nucleus and activates transcription of many genes that augment ER size and functions (29). PERK phosphorylates eukaryotic initiation factor 2 alpha (EIF2α) followed by activating transcription factor 4 (ATF4) activation (30). Consistently, we found that phosphorylated EIF2α (p-EIF2α, Fig. 5A) and mRNA levels of XBP1s and ATF4 (Fig. 5C) were increased by NGBR siRNA.
Figure 5.
NGBR sensitizes insulin actions by improving endoplasmic reticulum stress.A and C, HepG2 cells in 6-well plates were transfected with scrambled siRNA or NGBR siRNA (siCtrl or siNGBR, 50 nM) for 24 h. B and D, HepG2 cells in 6-well plates were transfected with NGBR expression or empty vector (pIRES-NGBR or pIRES-neo, 1 μg/well) for 12 h, followed by glucosamine hydrochloride (Glcn) treatment for 18 h. Expressions of binding immunoglobulin protein (BIP), pancreatic endoplasmic reticulum kinase (PERK), phosphorylated eukaryotic initiation factor 2 alpha (p-EIF2α), EIF2α and NGBR protein (A and B) and expression of activating transcription factor 4/6 (ATF4/6), X-box binding protein 1 (XBP1s), inositol-requiring enzyme 1α (IRE1α), BIP, DNA-damage inducible transcript 3 (CHOP), and skeletal muscle and kidney-enriched inositol polyphosphate phosphatase (SKIP) mRNA (C and D) were determined by western blot and qPCR, respectively. ∗p < 0.05 versus siCtrl or pIRES-neo; †p < 0.05 versus pIRES-neo plus Glcn treatment, n = 3.
Next, we determined if NGBR overexpression can attenuate Glcn-induced ER stress in HepG2 cells. Figure 5, B and D show Glcn-induced expression of BIP, PERK, p-EIF2α at protein levels and of ATF4, ATF6, XBP1s, IRE1α, BIP, CHOP, skeletal muscle and kidney-enriched inositol polyphosphate phosphatase (SKIP) at mRNA levels. High expressing NGBR reduced Glcn-induced expression of these genes (Fig. 5, B and D), especially SKIP expression, which may benefit the restoration of Glcn-impaired AKT phosphorylation (Fig. 4B).
NGBR overexpression in the liver increases AKT phosphorylation and abolishes HFD/STZ-induced expression of ER stress-related genes
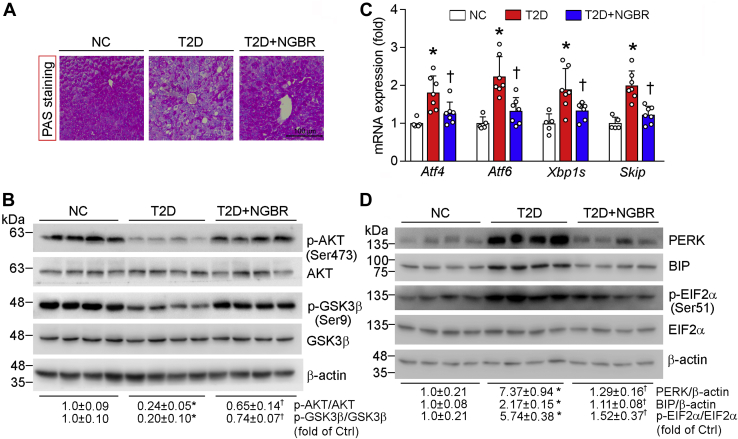
In the liver, insulin resistance can increase gluconeogenesis while reducing glycogen synthesis (31). Accumulation of glycogen in the liver can be determined by PAS staining (32). As shown in Figure 6A, AAV9-NGBR clearly induced the accumulation of hepatic glycogen content occurring to the liver of T2D group. It also increased phosphorylation of AKT and GSK3β in the liver of T2D+NgBR group (Fig. 6B), suggesting improvement of insulin sensitivity by NGBR overexpression.
Figure 6.
Hepatic NGBR overexpression activates AKT phosphorylation and abolishes HFD/STZ-induced expression of endoplasmic reticulum stress-related genes. The following assays were completed with liver samples collected from mice used in Figure 2A. A, PAS staining with liver paraffin sections. Scale bar: 100 μm. B and D, expression of p-AKT, AKT, p-GSK3β, and GSK3β protein (B). Expressions of pancreatic endoplasmic reticulum kinase (PERK), binding immunoglobulin protein (BIP), phosphorylated eukaryotic initiation factor 2 alpha (p-EIF2α), EIF2α and β-actin protein (D) were determined by western blot. C, expressions of activating transcription factor 4/6 (Atf4/6), X-box binding protein 1 (Xbp1s) and skeletal muscle and kidney-enriched inositol polyphosphate phosphatase (Skip) mRNA were determined by qPCR. ∗p < 0.05 versus NC; †p < 0.05 versus T2D; n = 5 (NC group) and n = 7 (T2D or T2D+NGBR group).
Next, we investigated the effects of NGBR on ER stress in mouse liver. High expressing NGBR reduced expression of Aft6 and PERK, the two ER stress-associated genes. Consequently, expression of their downstream genes (Atf4, Xbp1s, and Skip) and phosphorylation of EIF2α were reduced by NGBR overexpression (Fig. 6, C and D).
Improved systematic insulin sensitivity in HFD/STZ-AAV9-NGBR mice
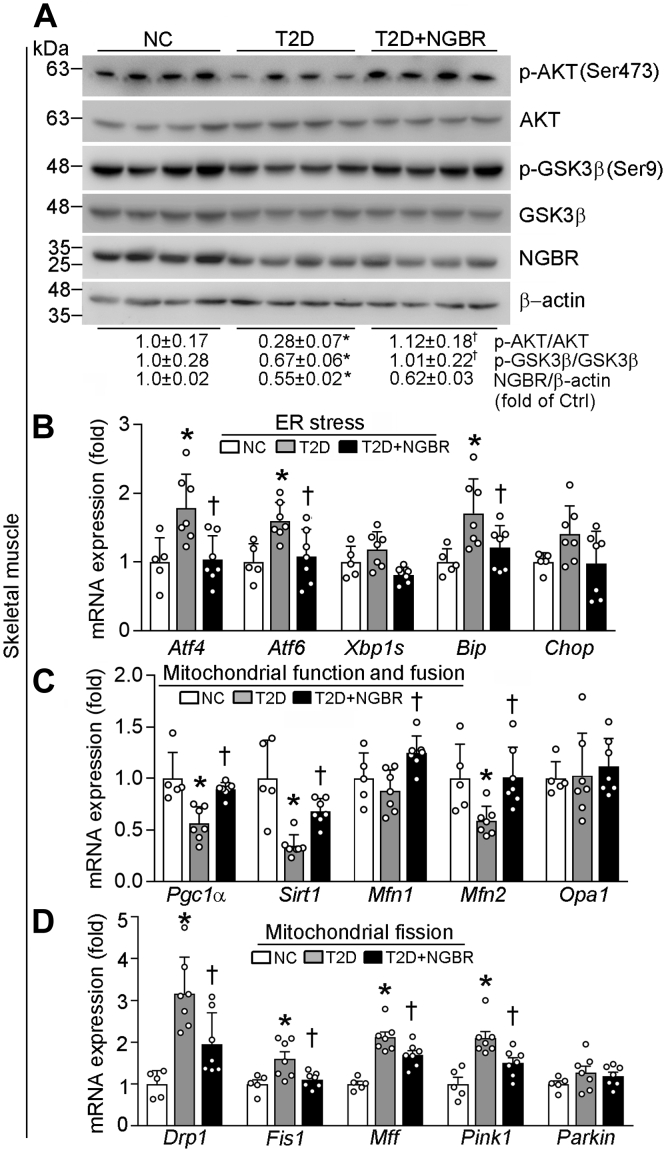
The reduction of blood glucose and serum NEFA levels (Fig. 3C, Table 1) implies improved insulin actions in AAV9-NGBR mice. Besides the liver, obesity-associated exposure of skeletal muscle to elevated NEFA can also contribute to insulin resistance (33). Indeed, compared with nondiabetic mice, phosphorylation of AKT and GSK3β in skeletal muscle were substantially reduced in T2D group, but they were restored to normal by hepatic NGBR overexpression (T2D+NGBR group) although NGBR expression in skeletal muscle was slightly affected (Fig. 7A). In addition, diabetes-induced expression of ER stress-related genes in skeletal muscle was also decreased in hepatic NGBR overexpressing mice (Fig. 7B).
Figure 7.
The systematic insulin sensitivity in HFD/STZ mice is improved by hepatic NGBR overexpression. Skeletal muscle samples were collected from mice used in Figure 2A, and the following assays were conducted. A, expressions of p-AKT, AKT, p-GSK3β, GSK3β, NGBR, and β-actin protein were determined by western blot. B–D, expressions of mRNA for genes related to endoplasmic reticulum stress: activating transcription factor 4/6 (Atf4/6), X-box binding protein 1 (Xbp1s), binding immunoglobulin protein (Bip), and DNA-damage inducible transcript 3 (Chop) or mitochondrial functions: peroxisome proliferator-activated receptor-γ coactivator 1α (Pgc1α), sirtuin 1 (Sirt1), mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), optic atrophy (Opa1), dynamin-related protein 1 (Drp1), mitochondrial fission 1 (Fis1), mitochondrial fission factor (Mff), Parkin, and (PTEN)-induced putative kinase 1 (Pink1) were determined by qPCR. ∗p < 0.05 versus NC; †p < 0.05 versus T2D; n = 5 (NC group), n = 7 (T2D or T2D+NGBR group).
Mitochondria is critical to maintaining normal skeletal muscle functions. Peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) functions as a master regulator of mitochondrial biogenesis by regulating expression of genes for mitochondrial fusion (34). Sirtuin 1 (SIRT1) is also involved in regulating expression of genes for mitochondrial functions (35). Both Pgc1α and Sirt1 were decreased by diabetes, but the decreases were abolished by hepatic NGBR overexpression (Fig. 7C). Mitochondrial fusion is regulated by the outer membrane fusion proteins mitofusin 1/2 (MFN1/2) and the inner membrane fusion protein optic atrophy 1 (OPA1). Although NGBR overexpression in the liver had little effect on Opa1 in skeletal muscle, it increased Mfn2 expression (Fig. 7C). Mitochondrial fission is also related to the etiology of T2D (36). Although liver NGBR overexpression had little effect on Parkin, it decreased expression of fission-related protein, such as dynamin-related protein 1 (Drp1), mitochondrial fission 1 (Fis1), mitochondrial fission factor (Mff), and PTEN-induced putative kinase 1 (Pink1) (Fig. 7D).
Discussion
In this study, we elucidated previously unrecognized functions of NGBR in enhancing insulin sensitivity and alleviating T2D. Our study demonstrates that loss of NGBR reduced insulin sensitivity while NGBR overexpression restored the impaired insulin sensitivity caused by HFD/STZ in vivo and Glcn in vitro. In addition, NGBR depletion increased expression of ER stress-related genes while its overexpression reduced Glcn-induced expression of these ER stress-associated genes. In vivo, liver NGBR overexpression ameliorated T2D by regulating expression of genes related to fatty acid synthesis, metabolism, and ER stress. Taken together, these findings suggest that increasing NGBR expression in the liver may be a potential strategy for T2D treatment.
T2D is caused by chronic insulin resistance and progressive decline in beta-cell function (37, 38). Optimal beta-cell function and mass are essential for glucose homeostasis while the beta-cell impairment leads to the development of diabetes (39, 40). Many studies have demonstrated that NEFA is one of the important links between obesity, insulin resistance, and T2D (41, 42). The elevated levels of circulating NEFA may increase the incidence of T2D (43). The chronic increase of circulating NEFA levels will reduce beta-cell functions and cause its apoptosis (lipotoxicity) and consequently aggravate the development of T2D (44). Therefore, reduction of elevated plasma NEFA levels might be an important therapeutic approach for T2D treatment. We previously demonstrated that NGBR deficiency in the liver increases hepatic fatty acids accumulation and plasma NEFA levels (8), suggesting that NGBR deficiency may be involved in lipotoxicity-caused insulin resistance/T2D. As shown in Figure 1, J and K, we observed the loss of beta cells in NGBRhepKO mice compared with the littermate control mice by HFD feeding. Reciprocally, NGBR overexpression in the liver reduced circulating NEFA levels in diabetic mice (Table 1), which may be another contributor to the protective effects of hepatic NGBR overexpression on islets. In addition, both glucose and inflammatory factors can induce beta-cell apoptosis. Glucose is the key physiological regulator of insulin secretion. Long-term adaptation of beta cells to conditions of increased demand of glucose may be triggered by hyperglycemic excursions. These excursions elicit beta-cell production of IL1 and other inflammatory factors (45, 46). As shown in Figure 1H, we observed the increased blood glucose in NGBRhepKO mice by HFD feeding, which was accompanied by increased TNFα and IL1β expression in the liver (Fig. S1). Indeed, the islet in pancreases of HFD/STZ mice receiving AAV9-NGBR infection exhibited normal distribution with a large insulin-positive cell core surrounded by a mantle of alpha cells, which differed from the disorganized islet architecture observed in pancreases of HFD/STZ mice receiving control AAV infection (Fig. 3F). It also clearly indicates that preserving hepatic NGBR expression can prevent the loss of beta cells in HFD/STZ mice (Fig. 3F). Consistently, the increased fasting serum insulin and C-peptide levels in diabetic mice were substantially reduced by hepatic NGBR overexpression (Fig. 3, A and B).
ER stress is also involved in the development of insulin resistance and progression to T2D (47). This response is linked to different processes during the progression of insulin resistance and T2D, including inflammation, lipid accumulation, insulin biosynthesis, and beta-cell apoptosis (47). Chronic exposure to high NEFA levels causes ER stress, which further contributes to insulin resistance and T2D (48). During ER stress, BIP is displaced to interact with unfolded/misfolded proteins, thereby resulting in BIP release from ER stress sensors, such as IRE1α, PERK, and ATF6, and consequent activation of UPR (49). The activated UPR then directly affects transcription of the key hepatic enzymes involved in gluconeogenesis/lipogenesis or interferes insulin signaling to promote its resistance (50). In addition, UPR promotes fat accumulation in hepatocytes, which may make additional contributions to insulin resistance indirectly (50). As shown in Figure 5, NGBR knockdown in HepG2 cells increased levels of BIP, PERK, and p-EIF2α as well as transcription of ATF6, BIP, IRE1α, and CHOP. In contrast, NGBR overexpression can ameliorate Glcn-induced ER stress in HepG2 cells. Similarly, hepatic NGBR overexpression reduced ER stress in HFD/STZ-treated mouse tissues (Figs. 6 and 7).
The increased NEFA levels may further enhance insulin resistance and induce ER or mitochondrial stress in skeletal muscle (51). In this study, we determined recovery of p-AKT and p-GSK3β in mouse skeletal muscle by NGBR overexpression. Correspondingly, HFD/STZ-induced expression of ER stress-related genes, decreased expression of mitochondrial fusion genes, and increased expression of mitochondrial fission genes in skeletal muscle were restored to normal (Fig. 7, B–D). Therefore, overexpression of NGBR reduces NEFA, which may contribute to improved insulin resistance in other tissues than the liver, such as the skeletal muscle.
AMPK plays a major role in regulating lipid metabolism; thus, it is believed as a mediator of metabolic effects of hormones, such as adiponectin, glucocorticoids, and insulin (52). In general, AMPK stimulates catabolism (fatty acid oxidation and glycolysis) while inhibiting anabolic pathways (gluconeogenesis, glycogen, fatty acid synthesis). It can also regulate insulin activity in the muscle and insulin synthesis/secretion in beta cells (53). Activation of insulin pathway by AMPK is related to activation of insulin receptor phosphorylation and insulin receptor substrate-1 through inhibition of mTOR (the pathway is implicated in the pathogenesis of insulin resistance) (54). We previously reported that genetic depletion of NGBR in the liver impairs AMPK activation (8). Here we found that NGBR inhibited mTOR phosphorylation. We further demonstrated that AMPK deficiency blocks insulin signaling pathway, and the beneficial effect of NGBR on insulin sensitivity is in an AMPK-dependent manner (Fig. 4). These results indicate that NGBR may also be involved in AMPK-mTOR pathway.
Although NGBR has been reported as the putative receptor for Nogo-B binding in endothelial cells (6, 15), if there is an interaction between NGBR and Nogo-B in other cell types has not been reported. In this study, we determined that neither diabetes nor high expressing NGBR in diabetic mouse liver had effect on Nogo-B expression in the liver (Fig. 2C, Fig. S2E). While inhibition of NGBR expression in HepG2 cells by siRNA or genetic deletion of NGBR expression in the liver had no effect on Nogo-B expression/secretion either (Fig. S2, A–D). Furthermore, we recently reported that reduced Nogo-B expression in mouse liver did not influence NGBR expression while still ameliorating ER stress (55). These observations suggest that NGBR and Nogo-B may demonstrate their functions independently.
In summary, our study defines the role of NGBR overexpression in insulin sensitivity and demonstrates that increasing hepatic NGBR expression may protect mice against T2D by improving insulin sensitivity, which is related to reduction of NEFA and ER stress and activation of AMPK pathway.
Experimental procedures
Reagents
Human insulin and D-(+)-Glcn were purchased from Sigma-Aldrich. STZ and puromycin were purchased from P212121. HFD (Cat# D12492) with 60 kcal% fat was purchased from Research Diets. Nile red was purchased from MedchemExpress. Micro Free Aliphatic Acid Content Assay Kit was purchased from Solarbio. Insulin and C-peptide ELISA kits were purchased from Elabscience Biotechnology. The human soluble Nogo-B ELISA kit was purchased from BioLegend. Mouse Nogo-B ELISA kit was purchased from MyBioSource. Schiff reagent was purchased from Yuanyebio. LabAssay Triglyceride was purchased from Wako Pure Chemical Industries. One-step TUNEL assay kit was purchased from KeyGEN BioTECH. Silencer siRNA Construction Kit was purchased from Life Technologies. Lipofectamine RNAiMAX Transfection Reagent and Lipofectamine 2000 Transfection Reagent were purchased from Thermo Fisher Scientific. AAV9-GFP and AAV9-NGBR-GFP were purchased from HanBio. Primary antibodies are listed in the Supplementary Materials.
Plasmids construction
Human NGBR cDNA with hemagglutinin (HA) tag was cloned into pIRES-neo vector (Clontech) as previously described (6). The NGBR overexpression vector was named as pIRES-NGBR while the empty vector as pIRES-neo.
Cell culture and siRNA transfection
HepG2 cells, a human hepatic cell line (ATCC HB-8065), was purchased from ATCC and cultured in complete DMEM medium. The AMPKα1 genome knockout HepG2 cell line was generated using the clustered regulatory interspaced short palindromic repeat (CRISPR)-associated 9 (Cas9) technology as described (56, 57). Mouse primary hepatocytes were isolated from C57BL/6 mouse liver as previously described (56). NGBR siRNA transfection was performed as described (58). All the cells were free of Mycoplasma.
Animal studies
C57BL/6 mice (male, 5-week-old) were purchased from the Animal Center of Nanjing University and received scheduled treatment as indicated in Figure 2A. NGBRfl/fl mice were crossbred with albumin-specific Cre mice [Alb-Cre, B6.CgTg(Alb-cre)21Mgn/J] (The Jackson Laboratory) on a C57BL/6J background. Alb-Cre efficiently induces hepatocyte-specific recombination of floxed genes to generate NGBRhepKO mice. Db/db mice (B6.BKS(D)-Leprdb/J) were purchased from the Jackson laboratory. The protocols for animal studies were approved by the Ethics Committee of Nankai University (Tianjin, China) and the Institutional Animal Care and Use Committee of the Medical College of Wisconsin. All animals were housed in a temperature-controlled environment with a 12/12 h light/dark cycles. All mice had free access to water and standard rodent chow or HFD (60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein). The studies were performed in compliance with the Guide for the Care and Use of Laboratory Animals published by the NIH.
Mice were treated with STZ while being fed HFD to induce T2D as described (59, 60). Briefly, the 5-week-old C57BL/6 male mice were fed HFD for 4 weeks and then i.p. injected STZ (50 mg day−1 kg−1 bodyweight in 0.1 M citrate buffer, pH 4.2–4.5) after 6-h fasting. The STZ injection was conducted for seven consecutive days while the mice drank 10% glucose solution during the treatment. Mice in NC group (n = 5) were injected citrate buffer and fed normal chow. All mice were maintained on their respective diets until the end of study.
To evaluate the effect of NGBR on T2D in mice, 14 diabetic mice (HFD/STZ-treated and the fasting blood glucose≥11.1 mM after STZ injection) from total of 22 mice were randomly divided into two groups (n = 7/group). At the end of 7 weeks of HFD feeding, the diabetic mice were i.v. injected once with either AAV9-GFP (named as T2D group) or AAV9-NGBR-GFP (named as T2D+NGBR group) at a dose of 1 × 1012 viral genomes (vg) per mouse (as scheduled in Fig. 2A). Bodyweight, food intake, and 12-h fasting glucose levels were monitored weekly. At the end of experiment, mice were anesthetized and euthanized in a CO2 chamber, followed by collection of blood and tissue samples.
GTT and ITT
GTT and ITT were carried out a few days before the end of study. For GTT, mice were orally administrated with glucose (0.5 g/kg bodyweight) after 12-h fasting, followed by determination of blood glucose levels at the indicated time points. For ITT, diabetic mice were i.p. injected with insulin (1 U/kg bodyweight) after 4-h fasting, followed by determination of blood glucose levels at the indicated time points. To conduct ITT in the nondiabetic (NGBRfl/fl and NGBRhepKO) mice, the animals were preadministrated glucose (0.5 g/kg bodyweight) for 2 h after 12-h fasting, then continued insulin injection and blood glucose test as described above.
Determination of lipid, Nogo-B, insulin, and C-peptide levels in serum, NEFA levels in serum and liver, Nogo-B level in HepG2 cell culture medium
After collection, mouse blood samples were kept for 2 to 3 h at room temperature followed by centrifugation for 20 min at 2000g. The serum was transferred into a new test tube. Serum levels of TG, ALP, AST, ALT, TBIL, and blood urea nitrogen (BUN) were measured by the automatic biochemical analyzer. Serum levels of Nogo-B, insulin and C-peptide were measured by the corresponding ELISA kits. Serum and liver NEFA levels were determined in accordance with the instruction of Micro Free Aliphatic Acid Content Assay Kit. HepG2 cell culture medium was collected, and Nogo-B level in the conditioned medium was measured using a human soluble Nogo-B ELISA kit.
Histological analysis and liver TG determination
After sacrificed, a piece of mouse liver was collected and fixed in 4% paraformaldehyde overnight. Samples were then incubated in 30% sucrose solution overnight followed by preparation of 5-μm frozen sections. The sections were conducted Oil Red O and Nile red staining as previously described (61, 62).
The liver TG content was determined with total lipid extract from a piece of liver as described (61).
Immunochemistry staining
For determining the area ratio of beta cells in pancreas, pancreas samples were fixed in paraffin and then used to prepare paraffin-embedded tissue sections. Immunohistochemical analysis of insulin status was performed on 5-μm sections using anti-insulin antibody. Tissue slides were blocked with 2.5% horse serum for 30 min at room temperature followed by incubation with anti-insulin antibody (1:500) overnight at 4 °C. Detection system used was 3,3’-diaminobenzidine (DAB) purchased from DAKO. Slides were counterstained using hematoxylin. Slides were observed and photographed with Leica microscope.
Immunofluorescent and TUNEL staining
To determine the structural integrity of islets, pancreas was fixed in 4% paraformaldehyde overnight and processed the conventional paraffin embedding. After being deparaffinized and permeabilized with 0.5% Triton X-100 for 10 min, the sections (5-μm) were incubated with citrate antigen repairing solution for 4 × 5 min at 94 to 98 °C. The sections were then blocked with 2% BSA for 2 h at room temperature and incubated with primary antibody (dilutions: anti-insulin, 1:500; anti-glucagon, 1:500) overnight at 4 °C. After removal of primary antibody by washing with PBS for three times, the sections were incubated with goat anti-mouse IgG-FITC and or anti-rabbit IgG-Rhodamine for 2 h. Following removal of the secondary antibody by washing with PBS for three times, the sections were stained with DAPI solution for nuclei. To assess apoptotic beta cells, TUNEL assay was performed using the One-step TUNEL assay kit. Nuclei were counterstained with DAPI.
Determination of protein expression by western blot and mRNA expression by qPCR
After treatment, total cellular proteins were extracted from HepG2 cells or mouse primary hepatocytes using a cell lysis buffer (63). Levels of p-AKT, total AKT, p-AMPK, total AMPKα1, p-GSK3β, total GSK3β, BIP, PERK, p-EIF2α, EIF2α, p-ACC, total ACC, p-Raptor, Raptor, p-mTOR, mTOR, p-IRS1, total IRS1, NGBR, or Nogo-B protein were determined by western blot using the corresponding primary antibodies (dilution: 1:1000∼2000) (63). A piece of liver or skeletal muscle tissue (∼30 mg) was used to extract total proteins followed by determination of p-AKT, total AKT, p-GSK3β, total GSK3β, PERK, BIP, p-EIF2α, EIF2α, NGBR, or Nogo-B protein expression by western blot. The same blot was reprobed with anti-GAPDH, Hsp90, or β-actin antibody to verify sample loading. The image of each western blot was scanned. The density of target band was quantified using Image J software (NIH) and then normalized by the density of GAPDH, Hsp90, or β-actin in the same sample. The value of the ratio of target band density to GAPDH, Hsp90, or β-actin density was further normalized by control sample, which was defined as 1.
After treatment, total cellular RNA was extracted from cells or a piece of mouse tissue. The cDNA was synthesized with 1 μg total RNA from each sample using a reverse transcription kit (New England Biolabs), followed by qPCR using SYBR Green Master Mix (Bio-Rad) and the primers listed in Table S1.
Data analysis
All the data were obtained from at least three independent experiments, and the representative results are presented. All results are expressed as mean ± SD. Statistical analysis was conducted using Prism 7 software (GraphPad Software). Statistical significance was evaluated using the unpaired two-tailed Student t-test or one-way ANOVA with post-hoc test among more than two groups. The significant difference was considered if p < 0.05 (n ≥ 3).
Data availability
All data shown are available in the article and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Miss Yi Chen received the American Society of Biochemistry and Molecular Biology Graduate/Postdoctoral Travel Award of this work at the “Experimental Biology 2020,” April 2020.
Author contributions
Y. C., W. H., Y. D., and Q. R. M.: conceptualization; Q. L., S. Z., and D. Z.: data curation; S. Z. and Z. W.: formal analysis; X. Y., Y. C., and X. L.: investigation; C. L. and J. H.: methodology; Y. C. and Q. L.: writing—original draft; Y. D. and Q. R. M.: writing—review and editing; Q. L.: software; Y. D.: supervision; Q. R. M.: project administration.
Funding and additional information
This work was funded by the International Science & Technology Cooperation Programs of China 2017YFE0110100 to J. H., Y. D., Y. C., and X. Y.; the China NSFC grants 81722046 to Y. D.; 81773727 and 81973316 to J. H., 31770863 to Y. C., and 81803517 to X. Y.; the Fundamental Research Funds for the Central Universities of China to Y. D., X. Y., and Y. C.
Edited by Dennis Voelker
Contributor Information
Qing Robert Miao, Email: Qing.Miao@nyulangone.org.
Yajun Duan, Email: yduan@hfut.edu.cn.
Supporting information
References
- 1.Alberti K.G., Zimmet P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Meshkani R., Adeli K. Hepatic insulin resistance, metabolic syndrome and cardiovascular disease. Clin. Biochem. 2009;42:1331–1346. doi: 10.1016/j.clinbiochem.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Targher G., Lonardo A., Byrne C.D. Nonalcoholic fatty liver disease and chronic vascular complications of diabetes mellitus. Nat. Rev. Endocrinol. 2018;14:99–114. doi: 10.1038/nrendo.2017.173. [DOI] [PubMed] [Google Scholar]
- 4.Chitturi S., Abeygunasekera S., Farrell G.C., Holmes-Walker J., Hui J.M., Fung C., Karim R., Lin R., Samarasinghe D., Liddle C., Weltman M., George J. NASH and insulin resistance: Insulin hypersecretion and specific association with the insulin resistance syndrome. Hepatology. 2002;35:373–379. doi: 10.1053/jhep.2002.30692. [DOI] [PubMed] [Google Scholar]
- 5.Tilg H., Moschen A.R., Roden M. NAFLD and diabetes mellitus. Nat. Rev. Gastroenterol. Hepatol. 2017;14:32–42. doi: 10.1038/nrgastro.2016.147. [DOI] [PubMed] [Google Scholar]
- 6.Miao R.Q., Gao Y., Harrison K.D., Prendergast J., Acevedo L.M., Yu J., Hu F., Strittmatter S.M., Sessa W.C. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10997–11002. doi: 10.1073/pnas.0602427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B., Hu W., Kumar S., Gonyo P., Rana U., Liu Z., Wang B., Duong W., Yang Z., Williams C., Miao Q. The Nogo-B receptor promotes Ras plasma membrane localization and activation. Oncogene. 2017;36:3406–3416. doi: 10.1038/onc.2016.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W., Zhang W., Chen Y., Rana U., Teng R.J., Duan Y., Liu Z., Zhao B., Foeckler J., Weiler H., Kallinger R.E., Thomas M.J., Zhang K., Han J., Miao Q.R. Nogo-B receptor deficiency increases liver X receptor alpha nuclear translocation and hepatic lipogenesis through an adenosine monophosphate-activated protein kinase alpha-dependent pathway. Hepatology. 2016;64:1559–1576. doi: 10.1002/hep.28747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Côté C., Rasmussen B., Duca F., Zadeh-Tahmasebi M., Baur J., Daljeet M., Breen D., Filippi B., Lam T. Resveratrol activates duodenal Sirt1 to reverse insulin resistance in rats through a neuronal network. Nat. Med. 2015;21:498–505. doi: 10.1038/nm.3821. [DOI] [PubMed] [Google Scholar]
- 10.Franko A., von Kleist-Retzow J., Neschen S., Wu M., Schommers P., Böse M., Kunze A., Hartmann U., Sanchez-Lasheras C., Stoehr O., Huntgeburth M., Brodesser S., Irmler M., Beckers J., de Angelis M. Liver adapts mitochondrial function to insulin resistant and diabetic states in mice. J. Hepatol. 2014;60:816–823. doi: 10.1016/j.jhep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Mann J. Diet and risk of coronary heart disease and type 2 diabetes. Lancet. 2002;360:783–789. doi: 10.1016/s0140-6736(02)09901-4. [DOI] [PubMed] [Google Scholar]
- 12.Sun T., Han X. Death versus dedifferentiation: The molecular bases of beta cell mass reduction in type 2 diabetes. Semin. Cell Dev. Biol. 2020;103:76–82. doi: 10.1016/j.semcdb.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Marselli L., Suleiman M., Masini M., Campani D., Bugliani M., Syed F., Martino L., Focosi D., Scatena F., Olimpico F., Filipponi F., Masiello P., Boggi U., Marchetti P. Are we overestimating the loss of beta cells in type 2 diabetes? Diabetologia. 2014;57:362–365. doi: 10.1007/s00125-013-3098-3. [DOI] [PubMed] [Google Scholar]
- 14.Rahier J., Guiot Y., Goebbels R., Sempoux C., Henquin J. Pancreatic beta-cell mass in European subjects with type 2 diabetes. Diabetes Obes. Metab. 2008;10 Suppl 4:32–42. doi: 10.1111/j.1463-1326.2008.00969.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B., Chun C., Liu Z., Horswill M.A., Pramanik K., Wilkinson G.A., Ramchandran R., Miao R.Q. Nogo-B receptor is essential for angiogenesis in zebrafish via Akt pathway. Blood. 2010;116:5423–5433. doi: 10.1182/blood-2010-02-271577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Z., Tseng Y., White M.F. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metab. 2010;21:589–598. doi: 10.1016/j.tem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo S. Insulin signaling, resistance, and the metabolic syndrome: Insights from mouse models into disease mechanisms. J. Endocrinol. 2014;220:T1–T23. doi: 10.1530/JOE-13-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirola L., Johnston A.M., Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47:170–184. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 19.Rossetti L., Hawkins M., Chen W., Gindi J., Barzilai N. In vivo glucosamine infusion induces insulin resistance in normoglycemic but not in hyperglycemic conscious rats. J. Clin. Invest. 1995;96:132–140. doi: 10.1172/JCI118013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore J.A., Miller W.P., Dennis M.D. Glucosamine induces REDD1 to suppress insulin action in retinal Muller cells. Cell. Signal. 2016;28:384–390. doi: 10.1016/j.cellsig.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruderman N.B., Carling D., Prentki M., Cacicedo J.M. AMPK, insulin resistance, and the metabolic syndrome. J. Clin. Invest. 2013;123:2764–2772. doi: 10.1172/JCI67227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoki K., Kim J., Guan K. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 23.Sakai K., Clemmons D.R. Glucosamine induces resistance to insulin-like growth factor I (IGF-I) and insulin in Hep G2 cell cultures: Biological significance of IGF-I/insulin hybrid receptors. Endocrinology. 2003;144:2388–2395. doi: 10.1210/en.2002-221133. [DOI] [PubMed] [Google Scholar]
- 24.Wallis M.G., Smith M.E., Kolka C.M., Zhang L., Richards S.M., Rattigan S., Clark M.G. Acute glucosamine-induced insulin resistance in muscle in vivo is associated with impaired capillary recruitment. Diabetologia. 2005;48:2131–2139. doi: 10.1007/s00125-005-1887-z. [DOI] [PubMed] [Google Scholar]
- 25.Wang S., Kaufman R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 28.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Liu C.Y., Schroder M., Kaufman R.J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 31.Krssak M., Brehm A., Bernroider E., Anderwald C., Nowotny P., Dalla Man C., Cobelli C., Cline G.W., Shulman G.I., Waldhausl W., Roden M. Alterations in postprandial hepatic glycogen metabolism in type 2 diabetes. Diabetes. 2004;53:3048–3056. doi: 10.2337/diabetes.53.12.3048. [DOI] [PubMed] [Google Scholar]
- 32.Boteon Y.L., Attard J., Boteon A., Wallace L., Reynolds G., Hubscher S., Mirza D.F., Mergental H., Bhogal R.H., Afford S.C. Manipulation of lipid metabolism during normothermic machine perfusion: Effect of defatting therapies on donor liver functional recovery. Liver Transpl. 2019;25:1007–1022. doi: 10.1002/lt.25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trujillo M.E., Scherer P.E. Adipose tissue-derived factors: Impact on health and disease. Endocr. Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 34.Hood D.A., Tryon L.D., Carter H.N., Kim Y., Chen C.C. Unravelling the mechanisms regulating muscle mitochondrial biogenesis. Biochem. J. 2016;473:2295–2314. doi: 10.1042/BCJ20160009. [DOI] [PubMed] [Google Scholar]
- 35.Ruderman N.B., Xu X.J., Nelson L., Cacicedo J.M., Saha A.K., Lan F., Ido Y. AMPK and SIRT1: A long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 2010;298:E751–E760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rovira-Llopis S., Bañuls C., Diaz-Morales N., Hernandez-Mijares A., Rocha M., Victor V. Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications. Redox Biol. 2017;11:637–645. doi: 10.1016/j.redox.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muoio D.M., Newgard C.B. Mechanisms of disease: Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 38.Ashcroft F.M., Rorsman P. Diabetes mellitus and the beta cell: The last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Florez J.C. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: Where are the insulin resistance genes? Diabetologia. 2008;51:1100–1110. doi: 10.1007/s00125-008-1025-9. [DOI] [PubMed] [Google Scholar]
- 40.Petrie J.R., Pearson E.R., Sutherland C. Implications of genome wide association studies for the understanding of type 2 diabetes pathophysiology. Biochem. Pharmacol. 2011;81:471–477. doi: 10.1016/j.bcp.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Kahn S.E., Hull R.L., Utzschneider K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 42.Karpe F., Dickmann J.R., Frayn K.N. Fatty acids, obesity, and insulin resistance: Time for a reevaluation. Diabetes. 2011;60:2441–2449. doi: 10.2337/db11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roden M., Price T.B., Perseghin G., Petersen K.F., Rothman D.L., Cline G.W., Shulman G.I. Mechanism of free fatty acid-induced insulin resistance in humans. J. Clin. Invest. 1996;97:2859–2865. doi: 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh Y.S., Bae G.D., Baek D.J., Park E.Y., Jun H.S. Fatty acid-induced lipotoxicity in pancreatic beta-cells during development of type 2 diabetes. Front. Endocrinol. (Lausanne) 2018;9:384. doi: 10.3389/fendo.2018.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maedler K., Sergeev P., Ris F., Oberholzer J., Joller-Jemelka H., Spinas G., Kaiser N., Halban P., Donath M. Glucose-induced beta cell production of IL-1beta contributes to glucotoxicity in human pancreatic islets. J. Clin. Invest. 2002;110:851–860. doi: 10.1172/JCI15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laybutt D., Glandt M., Xu G., Ahn Y., Trivedi N., Bonner-Weir S., Weir G. Critical reduction in beta-cell mass results in two distinct outcomes over time. Adaptation with impaired glucose tolerance or decompensated diabetes. J. Biol. Chem. 2003;278:2997–3005. doi: 10.1074/jbc.M210581200. [DOI] [PubMed] [Google Scholar]
- 47.Back S.H., Kaufman R.J. Endoplasmic reticulum stress and type 2 diabetes. Annu. Rev. Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poitout V., Robertson R.P. Glucolipotoxicity: Fuel excess and beta-cell dysfunction. Endocr. Rev. 2008;29:351–366. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J.M. Nuclear receptors resolve endoplasmic reticulum stress to improve hepatic insulin resistance. Diabetes Metab. J. 2017;41:10–19. doi: 10.4093/dmj.2017.41.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flamment M., Hajduch E., Ferre P., Foufelle F. New insights into ER stress-induced insulin resistance. Trends Endocrinol. Metab. 2012;23:381–390. doi: 10.1016/j.tem.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Rachek L.I. Free fatty acids and skeletal muscle insulin resistance. Prog. Mol. Biol. Transl. Sci. 2014;121:267–292. doi: 10.1016/B978-0-12-800101-1.00008-9. [DOI] [PubMed] [Google Scholar]
- 52.Towler M.C., Hardie D.G. AMP-activated protein kinase in metabolic control and insulin signaling. Circ. Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 53.Fu Z., Gilbert E.R., Liu D. Regulation of insulin synthesis and secretion and pancreatic beta-cell dysfunction in diabetes. Curr. Diabetes Rev. 2013;9:25–53. [PMC free article] [PubMed] [Google Scholar]
- 54.Chopra I., Li H.F., Wang H., Webster K.A. Phosphorylation of the insulin receptor by AMP-activated protein kinase (AMPK) promotes ligand-independent activation of the insulin signalling pathway in rodent muscle. Diabetologia. 2012;55:783–794. doi: 10.1007/s00125-011-2407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S., Guo F., Yu M., Yang X., Yao Z., Li Q., Wei Z., Feng K., Zeng P., Zhao D., Li X., Zhu Y., Miao Q.R., Iwakiri Y., Chen Y. Reduced Nogo expression inhibits diet-induced metabolic disorders by regulating ChREBP and insulin activity. J. Hepatol. 2020;73:1482–1495. doi: 10.1016/j.jhep.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 56.Ma C., Zhang W., Yang X., Liu Y., Liu L., Feng K., Zhang X., Yang S., Sun L., Yu M., Yang J., Li X., Hu W., Miao R.Q., Zhu Y. Functional interplay between liver X receptor and AMP-activated protein kinase alpha inhibits atherosclerosis in apolipoprotein E-deficient mice - a new anti-atherogenic strategy. Br. J. Pharmacol. 2018;175:1486–1503. doi: 10.1111/bph.14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W., Yang X., Chen Y., Hu W., Liu L., Zhang X., Liu M., Sun L., Liu Y., Yu M., Li X., Li L., Zhu Y., Miao Q.R., Han J. Activation of hepatic Nogo-B receptor expression-a new anti-liver steatosis mechanism of statins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:177–190. doi: 10.1016/j.bbalip.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong C., Zhao B., Long F., Liu Y., Liu Z., Li S., Yang X., Sun D., Wang H., Liu Q., Liang R., Li Y., Gao Z., Shao S., Miao Q.R. Nogo-B receptor promotes the chemoresistance of human hepatocellular carcinoma via the ubiquitination of p53 protein. Oncotarget. 2016;7:8850–8865. doi: 10.18632/oncotarget.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao X.G., Xu X., Wang G.H., Lei M., Quan L.L., Cheng Y.H., Wan P., Zhou J.P., Chen J., Hu L.H., Shen X. BBT improves glucose homeostasis by ameliorating beta-cell dysfunction in type 2 diabetic mice. J. Endocrinol. 2015;224:327–341. doi: 10.1530/JOE-14-0721. [DOI] [PubMed] [Google Scholar]
- 60.Zhou T.T., Quan L.L., Chen L.P., Du T., Sun K.X., Zhang J.C., Yu L., Li Y., Wan P., Chen L.L., Jiang B.H., Hu L.H., Chen J., Shen X. SP6616 as a new Kv2.1 channel inhibitor efficiently promotes beta-cell survival involving both PKC/Erk1/2 and CaM/PI3K/Akt signaling pathways. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y., Duan Y., Yang X., Sun L., Liu M., Wang Q., Ma X., Zhang W., Li X., Hu W., Miao R.Q., Xiang R., Hajjar D.P., Han J. Inhibition of ERK1/2 and activation of LXR synergistically reduce atherosclerotic lesions in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2015;35:948–959. doi: 10.1161/ATVBAHA.114.305116. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y., Jiang L., Sun C., Ireland N., Shah Y.M., Liu Y., Rui L. Insulin/Snail1 axis ameliorates fatty liver disease by epigenetically suppressing lipogenesis. Nat. Commun. 2018;9:2751. doi: 10.1038/s41467-018-05309-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y., Duan Y., Kang Y., Yang X., Jiang M., Zhang L., Li G., Yin Z., Hu W., Dong P., Li X., Hajjar D.P., Han J. Activation of liver X receptor induces macrophage interleukin-5 expression. J. Biol. Chem. 2012;287:43340–43350. doi: 10.1074/jbc.M112.403394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data shown are available in the article and the supporting information.