Figure 1. The BET Bromodomain Family of Epigenetic Reader Proteins In Transcriptional Regulation.
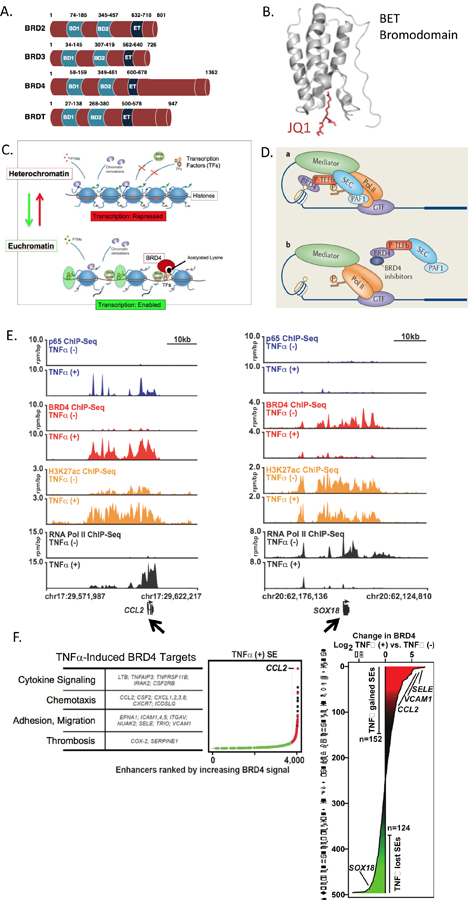
A. The Bromodomain and Extra-Terminal (BET) family members BRD2, BRD3, BRD4 and testes-restricted BRDT, all possess two bromodomains BD1 and BD2, as well as an extra-terminal (ET) domain. BRD4’s unique c-terminal domain (CTD) may facilitate its interaction with P-TEFb and RNA Polymerase II (RNAPII). B. BET bromodomain module predicted ribbon structure includes a hydrophobic pocket that binds to acetylated lysine residues on the 5’ end of histone tails. The pan-BETi JQ1 fits tightly and irreversibly in this pocket, disrupting BET’s association with chromatin and regulation of transcription. C. BET action is coupled to epigenetic mechanisms involved in the transition from heterochromatin to euchromatin. Heterochromatin consists of DNA tightly wound around histone spools, which results in DNA compaction but precludes transcription factor (TF) access to regulatory regions of DNA, repressing gene expression. For transcription to proceed, chromatin transitions to the euchromatin state, which opens DNA regulatory regions, allowing access for transcriptional machinery. BETs, in this case BRD4, binds to acetylated lysines on histone tails and facilitates assembly of TFs and RNAPII. D. Schematic representation of BRD4 binding to a histone tail and allowing for assembly of key players of the transcriptional apparatus, including RNAPII and P-TEFb. Mediator involves 30 protein subunits involved in the “pre-initiation complex”. In the presence of a BETi, BRD4 histone tail binding is disrupted, preventing BRD4 from its function as a transcriptional scaffold and mediator. E. Browser views of ChIP-Seq data in human umbilical vein ECs at baseline and after TNFα stimulation serves as an example of BRD4 regulation of vascular cell gene expression. E Left. Immunoprecipitation of individual proteins amassing at the 5’ end of the chemokine CCL2 gene body (arrow, chr17:29,622,217). Data is shown in pairs, without (upper) or with (lower) TNFα stimulation. At the top, in blue, absent TNFα stimulation, minimal immunoprecipitated p65 is found proximal to the CCL2 start site. As expected, TNFα stimulation recruits p65 to the CCL2 promoter region. In red, BRD4 protein accumulates upstream of the CCL2 start site after TNFα stimulation but was not present under basal conditions. In orange, H3K27Ac indicates an activated promoter after TNFα stimulation. In black, RNAPII is present only after TNFα stimulation and is evident ‘walking down’ the CCL2 gene body, consistent with transcription. JQ1 treatment of ECs blocks BRD4 and blocks these responses (not shown). E. Right. The ChIP-Seq browser view data for Sox18 (arrow) is shown. In contrast to CCL2 and other TNFα-induced, BRD4-regulated genes, Sox18 shows BRD4 and RNAPII association under basal conditions that is “lost” upon TNFα stimulation. F. Left: Multiple TNFα-stimulated pro-atherosclerotic target genes involve BRD4 action at super-enhancer regions. Right: BRD4 accumulation is shown where TNFα either increased (red, “gained”) or decreased (green, “lost”) BRD4 at respective super-enhancers. Each horizontal line corresponds to a distinct gene, including e-selectin (SELE), VCAM1, CCL2 and SOX18. For additional details, see text and associated references.
