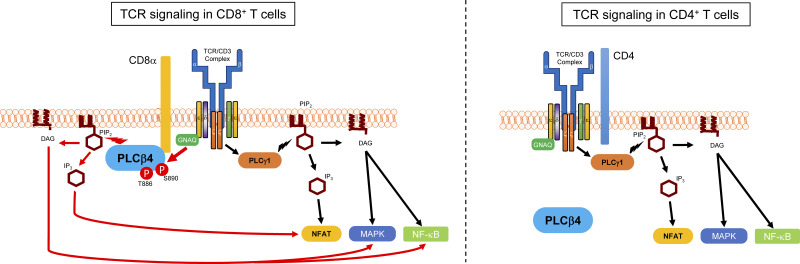
Sasai, Ma, et al. demonstrate that PLCβ4 is specifically required for TCR signaling in CD8+ T cells but not CD4+ T cells, indicating that TCR signaling of CD8+ T cells is qualitatively distinct from that of CD4+ T cells.
Abstract
Because of their common signaling molecules, the main T cell receptor (TCR) signaling cascades in CD4+ and CD8+ T cells are considered qualitatively identical. Herein, we show that TCR signaling in CD8+ T cells is qualitatively different from that in CD4+ T cells, since CD8α ignites another cardinal signaling cascade involving phospholipase C β4 (PLCβ4). TCR-mediated responses were severely impaired in PLCβ4-deficient CD8+ T cells, whereas those in CD4+ T cells were intact. PLCβ4-deficient CD8+ T cells showed perturbed activation of peripheral TCR signaling pathways downstream of IP3 generation. Binding of PLCβ4 to the cytoplasmic tail of CD8α was important for CD8+ T cell activation. Furthermore, GNAQ interacted with PLCβ4, mediated double phosphorylation on threonine 886 and serine 890 positions of PLCβ4, and activated CD8+ T cells in a PLCβ4-dependent fashion. PLCβ4-deficient mice exhibited defective antiparasitic host defense and antitumor immune responses. Altogether, PLCβ4 differentiates TCR signaling in CD4+ and CD8+ T cells and selectively promotes CD8+ T cell–dependent adaptive immunity.
Graphical Abstract
Introduction
T cells play a critical role in adaptive immunity for pathogen elimination and tumor surveillance (Gaud et al., 2018). T cells express a TCR-CD3 complex containing TCRαβ and CD3ζγεδεζ subunits on their surfaces (Fu et al., 2014). Under physiological conditions, T cell activation is initiated by engagement of TCRs on T cells and peptides in complex with major histocompatibility complex (pMHC) molecules on APCs (Samelson and Klausner, 1988). After binding of the TCR to the pMHC, tyrosine residues of immunoreceptor tyrosine-based activation motifs in the cytosolic region of the CD3ζ chain are phosphorylated by lymphocyte protein tyrosine kinase (Lck), recruiting ζ-chain-associated protein kinase of 70 kD (ZAP70; Courtney et al., 2018). Subsequently, ZAP70 phosphorylates the transmembrane adaptor molecule linker for the activation of T cells (LAT), which serves as a signaling hub and recruits other signaling molecules to the plasma membrane, including SH2-domain–containing leukocyte protein of 76 kD (SLP76) and phospholipase C γ1 (PLCγ1; Dustin and Choudhuri, 2016). The LAT/SLP76 complex together with IL-2–inducible T cell tyrosine kinase activates PLCγ1, catalyzing hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to produce inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG; Rhee, 2001). IP3 and DAG are important for incrementing cytosolic Ca2+ levels to activate nuclear factor of activated T cells (NFAT) and phosphorylate MAPKs and protein kinase C θ (PKCθ), which activates NF-κB, resulting in production of various cytokines, T cell proliferation, and T cell effector function (Rao and Hogan, 2009; Samelson, 2011).
Although these core TCR signaling molecules are shared between CD4+ and CD8+ T cells, it is widely assumed that CD4 and CD8 are not accessory molecules but coreceptors that potentiate activation signals (Eichmann et al., 1989; Janeway, 1992; Parnes, 1989). Both CD4 and CD8α coreceptors augment CD3-TCR signaling, since the cytoplasmic regions of both CD4 and CD8α interact with Lck (Rudd et al., 1988; Veillette et al., 1988). Since Lck preferentially binds CD4 rather than CD8α (Veillette et al., 1989), TCR signaling pathways in CD4+ and CD8+ T cells are quantitatively different. However, given the shared signaling molecules, it is widely accepted that TCR signaling pathways in CD4+ and CD8+ T cells are qualitatively identical (Artyomov et al., 2010; Fu et al., 2014; Gaud et al., 2018). The cytoplasmic tail of CD8α contains a CXCP motif important for Lck binding (Shaw et al., 1990; Turner et al., 1990). CD8α-deficient mice restored with a mutant CD8α in which the Lck-binding motif is replaced with alanine display milder phenotypes in terms of CD8+ T cell activation and T cell development in the thymus than do CD8-deficient mice restored with tailless CD8α (Chan et al., 1993; Fung-Leung et al., 1993), suggesting that there are other signaling molecules that bind to the cytoplasmic regions of CD8α (Zamoyska, 1994). However, such signaling molecules that specifically bind CD8α and differentiate the CD8+ TCR signaling pathway are unknown.
PLC regulates cellular concentrations of PIP2 as substrate or IP3 and DAG as products to control complex cellular homeostasis. IP3 and DAG act as second messengers (Kadamur and Ross, 2013). IP3 binds to the IP3 receptor on the endoplasmic reticulum and increases cytosolic Ca2+ levels. Ca2+ and DAG activate PKC (Rhee, 2001). The mammalian PLC family consists of 13 members classified into 6 subtypes (β, γ, δ, ε, ζ, and η) based on amino acid sequence and domain structure (Kadamur and Ross, 2013). All PLCs possess a highly conserved catalytic core containing an N-terminal pleckstrin homology domain, an EF-hands motif, a split X+Y catalytic domain, and a C2 domain (Kadamur and Ross, 2013). In TCR signaling, PLCγ1 plays a critical role in activation of both CD4+ and CD8+ T cells under physiological conditions (Samelson, 2002). In contrast, bacterial superantigens activate TCR signaling pathways independently of PLCγ1 (Bueno et al., 2006). Moreover, such superantigen-induced activation does not require Lck, ZAP70, or CD4. Furthermore, PLCβ inhibition and PLCβ1 silencing each partially block superantigen-induced T cell activation (Bueno et al., 2006), suggesting that PLCβ1 is involved in activation under pathological conditions. PLCβ4 is the last member of the PLCβ family and is highly expressed in the retina and cerebellar Purkinje cells (Peng et al., 1997; Watanabe et al., 1998). PLCβ4 deficiency leads to defects in the visual process and ataxia, suggesting its neurological significance (Jiang et al., 1996; Kano et al., 1998; Kim et al., 1997). Despite these established neurological roles, the immune functions of PLCβ4, especially in T cell signaling pathways, remain unexplored (Kawakami and Xiao, 2013). In this study, we examine the roles of PLCβ4 in TCR signaling and immune response and find that PLCβ4 is selectively required for activation of TCR signaling in CD8+ T cells and physiologically important for antiparasitic and antitumor adaptive immune responses.
Results
PLCβ4 deficiency impairs activation of CD8+ T cells but not CD4+ T cells
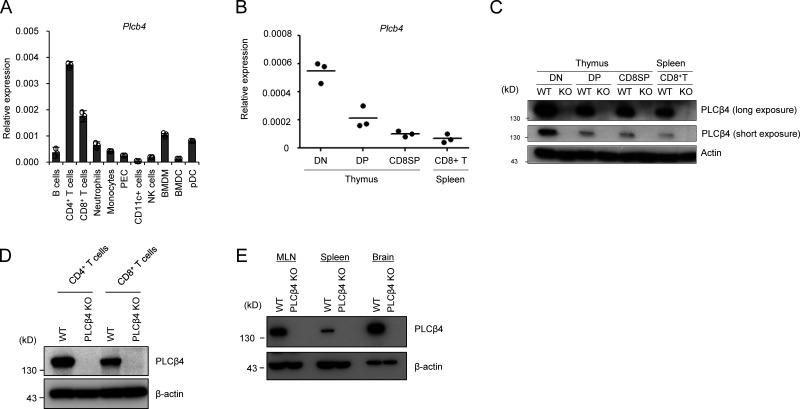
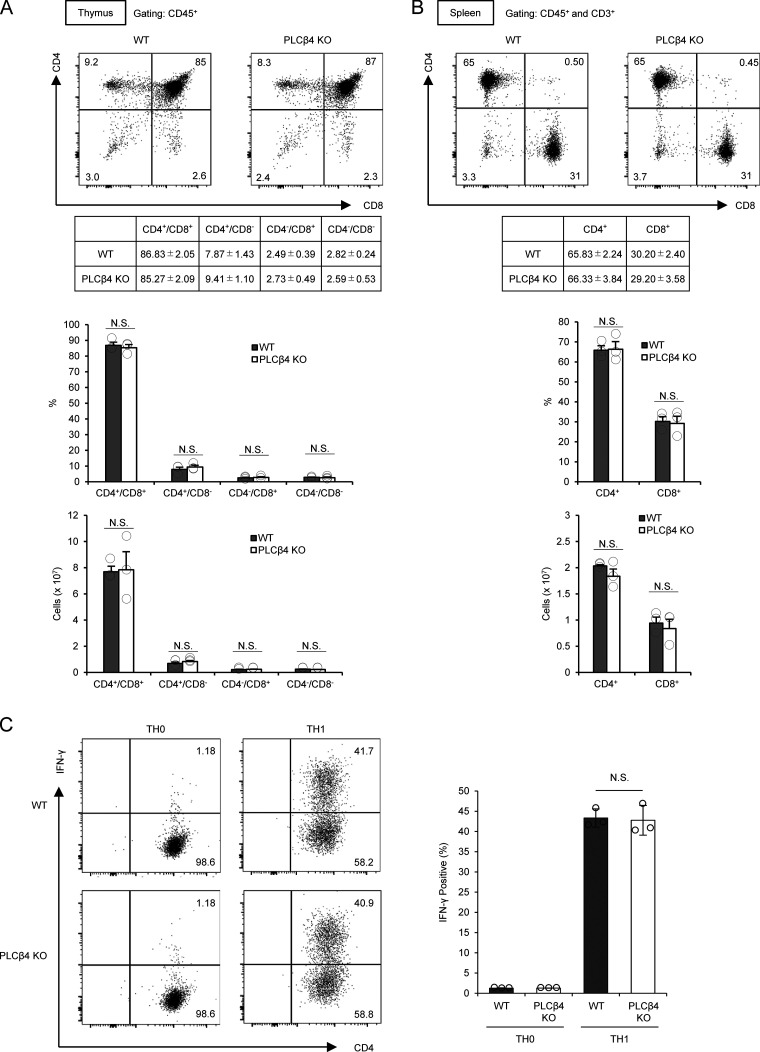
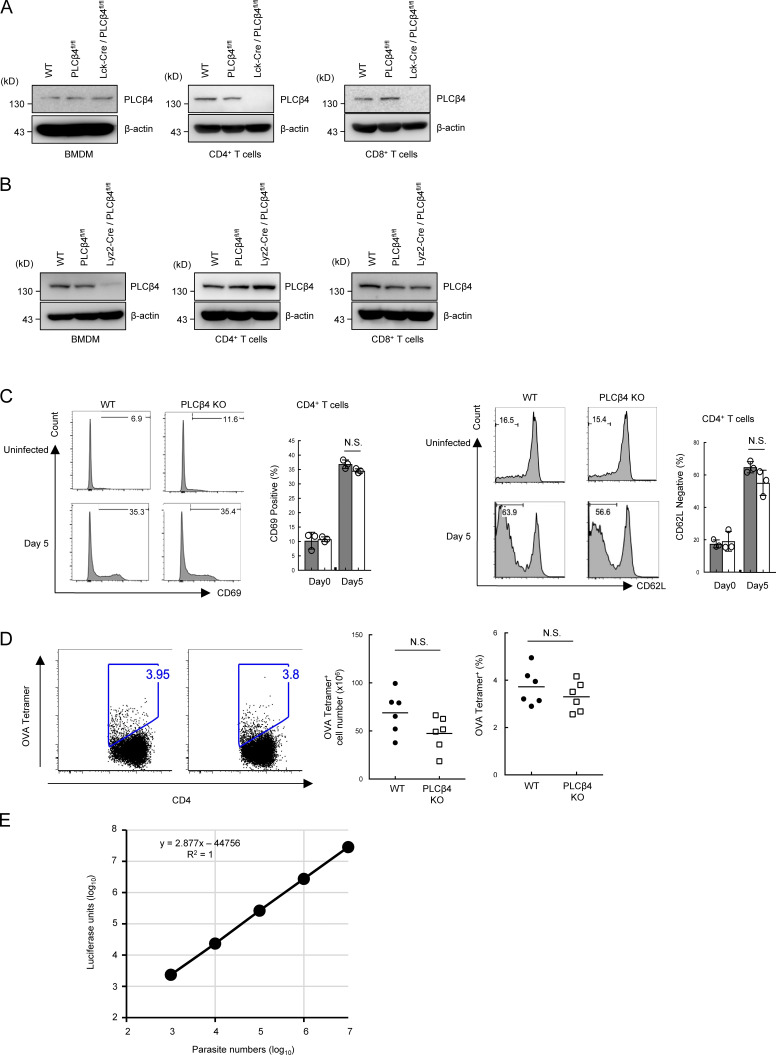
The immune functions of PLCβ4 remain unexplored (Kawakami and Xiao, 2013). Although several transcriptional and proteomic data repositories such as BioGPS and ImmProt showed low expression of PLCβ4 transcripts and proteins (Howden et al., 2019; Rieckmann et al., 2017; Voisinne et al., 2019), we found that PLCβ4 transcripts were low but surely detected in both CD4+ and CD8+ splenic T cells (Fig. 1 A). Moreover, the PLCβ4 mRNA and proteins in double-negative (CD4−CD8−), double-positive (CD4+CD8+), and single-positive and CD8+ T cells in the thymus were detected by quantitative RT-PCR and Western blot, respectively (Fig. 1, B and C), suggesting a potential role of PLCβ4 in T cell function. To determine the roles of PLCβ4 in T cell–mediated immune responses under physiological conditions, we generated PLCβ4-deficient mice with a gene-trapping strategy using embryonic stem cells (Fig. S1, A and B). PLCβ4 protein expression was abolished in organs and cells, including in CD4+ and CD8+ T cells (Fig. 1, C–E). When we first assessed T cell development, PLCβ4-deficient mice showed intact cell numbers and frequency of double-negative (CD4−CD8−), double-positive (CD4+CD8+), and single-positive CD4+ T cells and CD8+ T cells in the thymus (Fig. S2 A). In addition, the cellularity of splenic CD4+ T cells and CD8+ T cells in WT and PLCβ4-deficient mice was comparable (Fig. S2 B). Furthermore, in vitro differentiation of naive CD4+ T cells toward Th1 cells in WT and PLCβ4-deficient mice was also comparable (Fig. S2 C). Moreover, composition of CD44 and CD62L of CD4+ or CD8+ T cells was comparable in WT and PLCβ4-deficient mice (Fig. S3, A and B).
Figure 1.
PLCβ4 is expressed in CD4+ and CD8+ T cells. (A) Quantitative PCR analysis of the levels of Plcb4 mRNA in the indicated cells. (B) Quantitative PCR analysis of the levels of Plcb4 mRNA in the indicated thymocytes and splenic CD8+ T cells. (C) Western blot analysis of PLCβ4 proteins in the indicated thymocytes and splenic CD8+ T cells. (D) Purified CD4+ or CD8+ T cells (2 × 106 cells each) from WT or PLCβ4-deficient mice were lysed. PLCβ4 protein levels in the indicated lysates were determined by Western blotting with indicated Abs. (E) PLCβ4 protein levels in the indicated organs from WT or PLCβ4-deficient mice were determined by Western blotting with indicated Abs. Indicated values are means ± SD of three biological replicates (A). Data are representative two independent experiments (C–E) or cumulative of three independent experiments (B).
Figure S1.
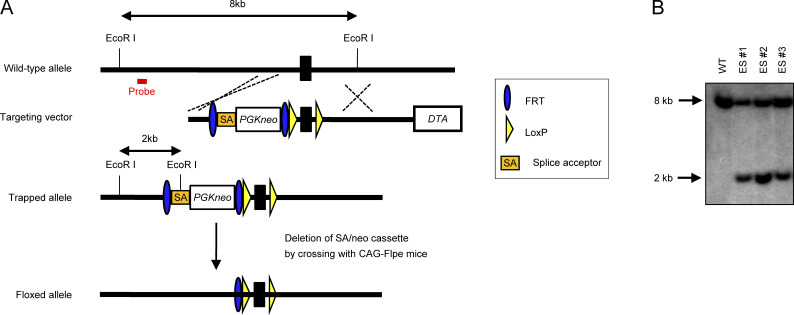
Generation of PLCβ4-deficient mice. (A) The gene-targeting strategy for Plcb4 locus using ES cells. (B) Southern blot analysis of total genomic DNA extracted from the WT or PLCβ4+/− ES cells. Genomic DNA was digested with EcoRI, electrophoresed, and hybridized with the radiolabeled probe indicated in (A). Southern blotting resulted in an 8-kb band for WT locus and a 2-kb band for mutated locus. Data are representative two independent experiments (B).
Figure S2.
PLCβ4-deficient mice show intact T cell development and Th1 differentiation. (A) Percentages and cell numbers of CD4−/CD8−, CD4+/CD8−, CD4−/CD8+, and CD4+/CD8+ in the thymocytes from WT or PLCβ4-deficient mice were analyzed by flow cytometry. (B) Percentages and cell numbers of CD4+ and CD8+ T cells in the splenocytes from WT or PLCβ4-deficient mice were analyzed by flow cytometry. (C) Naive CD4+ T cells from WT or PLCβ4-deficient mice were cultured under Th1-polarizing conditions for 3 d. Frequencies of IFN-γ producers were determined by intracellular staining using flow cytometry. Indicated values are means ± SD of three biological replicates (A–C). N.S., nonsignificant.
Figure S3.
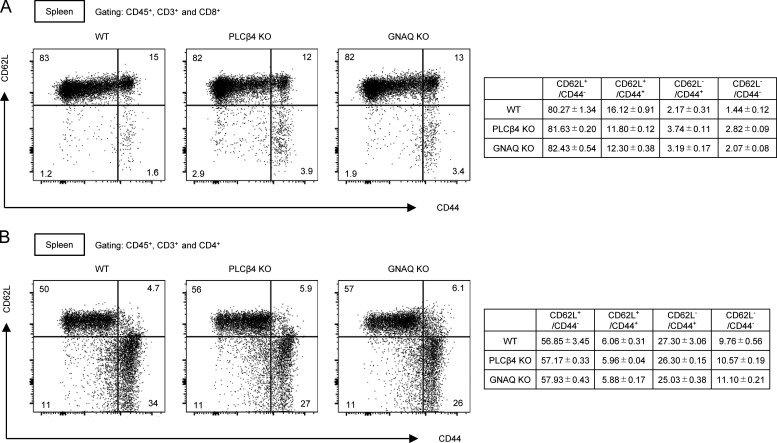
Normal CD44/CD62L profiles of CD4+ and CD8+ T cells in PLCβ4-deficient or GNAQ-deficient mice. (A and B) Percentages of CD4+ (A) and CD8+ (B) T cells in the splenocytes from WT or PLCβ4-deficient mice were analyzed by flow cytometry. Indicated values are means ± SD of three biological replicates (A and B).
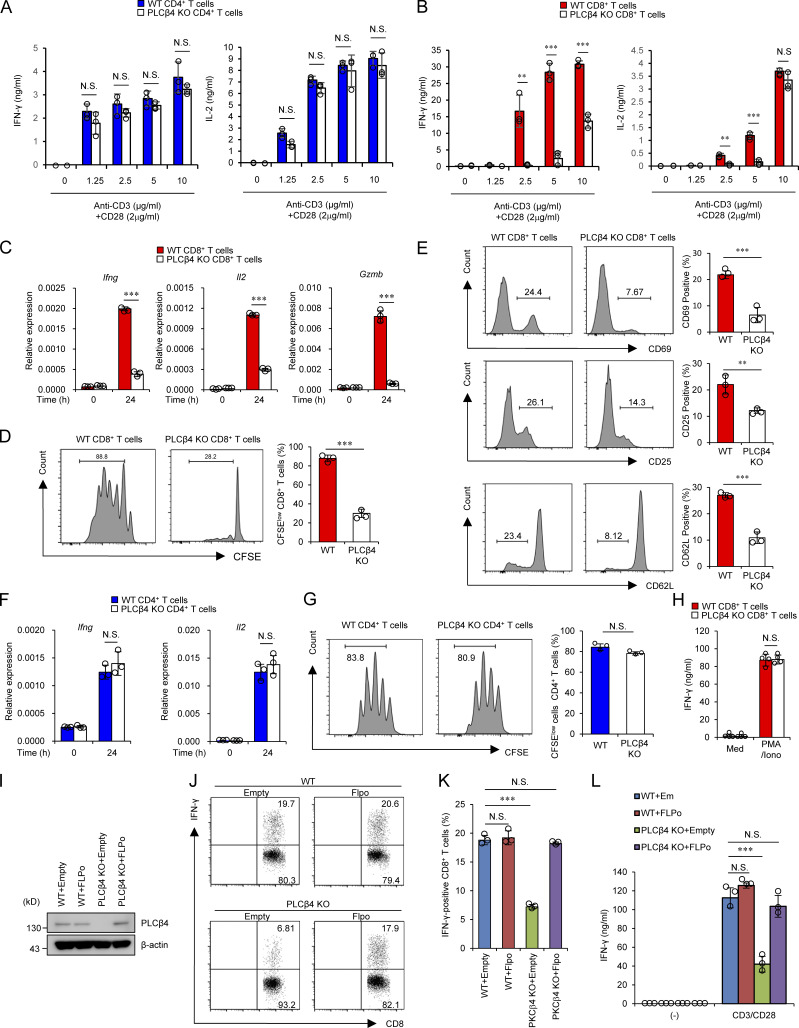
We next analyzed whether PLCβ4 is involved in cytokine production in CD4+ and CD8+ T cells (Fig. 2, A and B). CD4+ and CD8+ T cells isolated from the spleens of PLCβ4-deficient and WT mice were stimulated with various concentrations of α-CD3 (0, 1.25, 2.5, 5, and 10 µg/ml) in addition to 2 µg/ml of α-CD28 and tested for production of IFN-γ and IL-2 (Fig. 2, A and B). Production of IFN-γ and IL-2 was intact in PLCβ4-deficient CD4+ T cells (Fig. 2 A). Notably, production of both cytokines was severely impaired in PLCβ4-deficient CD8+ T cells in response to 1.25, 2.5, or 5 µg/ml of α-CD3, whereas the defects were less obvious at the extremely high α-CD3 concentration (10 µg/ml; Fig. 2 B). In response to optimal α-CD3/α-CD28 stimulation (5 µg/ml and 2 µg/ml, respectively), PLCβ4-deficient CD8+ T cells produced significantly fewer mRNA transcripts of Ifng, Il2, and Gzmb (Fig. 2 C), proliferated less (Fig. 2 D), and displayed lower expression of activation surface markers, such as CD69, CD25, and CD62Llow, than did WT cells (Fig. 2 E). In sharp contrast, expression of Ifng and Il2 mRNAs and proliferation in WT and PLCβ4-deficient CD4+ T cells were comparable (Fig. 2, F and G), indicating that loss of PLCβ4 selectively impairs α-CD3/CD28-mediated activation of CD8+ T cells, but not of CD4+ T cells. Moreover, PMA/ionophore-mediated IFN-γ production was intact in PLCβ4-deficient CD8+ T cells (Fig. 2 H). We next assessed whether defective IFN-γ production in PLCβ4-deficient CD8+ T cells is due to cell-intrinsic loss of PLCβ4 expression (Fig. 2, I–L). Purified CD8+ T cells from WT or PLCβ4-deficient mice were retrovirally transduced with Flp recombinase to remove the gene-trapping cassette flanked by two FRT sequences (Figs. 2 I and S1 A). Retroviral transduction of Flp recombinase into PLCβ4-deficient CD8+ T cells recovered PLCβ4 protein levels to those in WT cells (Fig. 2 I). We then assessed IFN-γ production in response to α-CD3/CD28 stimulation. Restoration of PLCβ4 expression in PLCβ4-deficient CD8+ T cells recovered IFN-γ production (Fig. 2, J–L). Taken together, these data indicate that PLCβ4 expression in CD8+ T cells plays an important role in cellular activation in response to α-CD3/CD28 stimulation.
Figure 2.
PLCβ4 deficiency exhibits impaired activation of CD8+ T cells, but not CD4+ T cells. (A and B) Purified CD4+ T cells (A) and CD8+ T cells (B) from WT and PLCβ4-deficient mice were stimulated with indicated doses of anti-CD3 and anti-CD28 for 72 h. Concentrations of IFN-γ and IL-2 in the culture supernatants were measured by ELISA. (C) Quantitative PCR analysis of the levels of Ifng, Il2, and Gzmb mRNA in WT or PLCβ4-deficient CD8+ T cells unstimulated or stimulated with anti-CD3 (5 µg/ml) and anti-CD28 (2 µg/ml) for 24 h. (D) CFSE-labeled WT or PLCβ4-deficient CD8+ T cells were stimulated with anti-CD3 (5 µg/ml)/anti-CD28 (2 µg/ml) for 3 d. The fluorescence intensity of CFSE-labeled CD8+ T cells was analyzed by flow cytometry. (E) Flow cytometry analysis of CD69, CD25, and CD62L in WT or PLCβ4-deficient CD8+ T cells stimulated with anti-CD3 (5 µg/ml) and anti-CD28 (2 µg/ml) for 6 h. (F) Quantitative PCR analysis of the levels of Ifng and Il2 mRNA in WT or PLCβ4-deficient CD4+ T cells unstimulated or stimulated with anti-CD3 (5 µg/ml) and anti-CD28 (2 µg/ml) for 24 h. (G) CFSE-labeled WT or PLCβ4-deficient CD4+ T cells were stimulated with anti-CD3 (5 µg/ml)/anti-CD28 (2 µg/ml) for 3 d. The fluorescence intensity of CFSE-labeled CD4+ T cells was analyzed by flow cytometry. (H) Purified CD8+ T cells from WT and PLCβ4-deficient mice were stimulated with PMA (50 ng/ml) and ionophore (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. Med, medium. (I–L) Purified CD8+ T cells from WT or PLCβ4-deficient mice were retrovirally transduced with empty or Flp recombinase. (I) PLCβ4 protein levels in the indicated lysates were determined by Western blotting. (J and K) Frequencies of IFN-γ producers were determined by flow cytometry after stimulation with anti-CD3 (0.5 µg) and anti-CD28 (1 µg/ml). (L) Transduced cells were stimulated with anti-CD3 (0.5 µg) and anti-CD28 (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. Indicated values are means ± SD of three biological replicates (A–L). **, P < 0.01; ***, P < 0.001; N.S., nonsignificant. Data are representative two (I) or three (J) independent experiments.
PLCβ4 deficiency impairs activation of downstream TCR signaling pathways
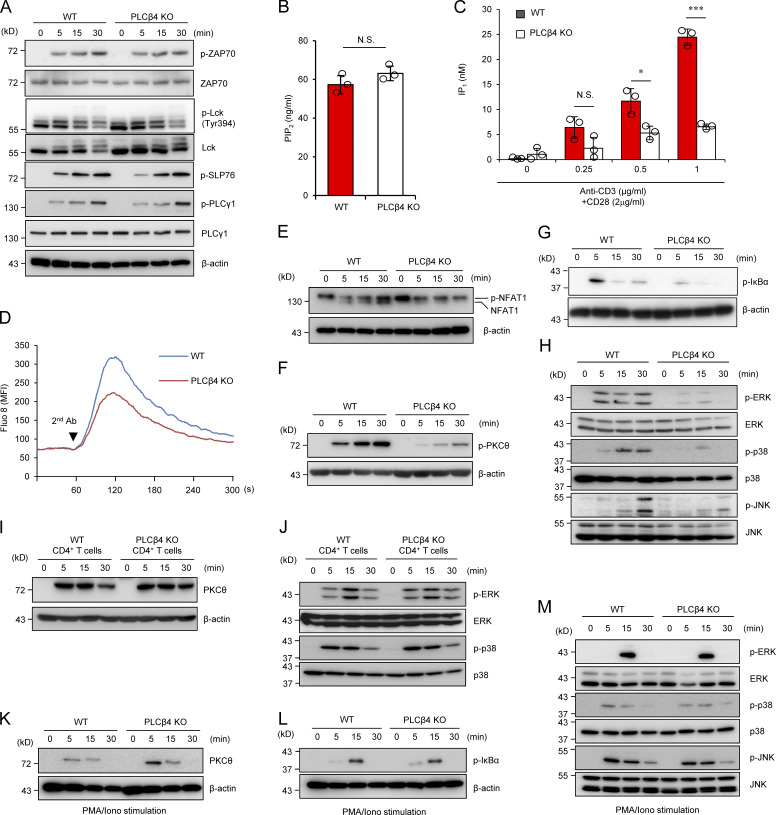
Since α-CD3/CD28-induced mRNA expression of Ifng, Il2, and Gzmb was reduced in PLCβ4-deficient CD8+ T cells (Fig. 2 C), we next assessed activation of TCR signaling pathways in the same cells. α-CD3/CD28-induced phosphorylation of ZAP70, SLP76, Lck, and PLCγ1, which all are located at the proximal TCR complex (Horejsí et al., 2004), was intact in PLCβ4-deficient CD8+ T cells (Fig. 3 A). PLCβ4 hydrolyzes PIP2 and generates IP3 and DAG (Kadamur and Ross, 2013). Therefore, we next examined whether PLCβ4 plays a role in the production of IP3 after TCR stimulation. IP1 is a downstream metabolite of IP3, which accumulates in cells following activation of PLCs (Thomsen et al., 2005). Although basal PIP2 levels in PLCβ4-deficient CD8+ T cells were comparable to those in WT cells (Fig. 3 B), α-CD3/CD28-induced IP1 production was decreased in PLCβ4-deficient CD8+ T cells compared with that in WT cells (Fig. 3 C). PLCβ4-deficient CD8+ T cells displayed reduced Ca2+ flux (Fig. 3 D) and decreased NFAT1 dephosphorylation by TCR stimulation compared with levels in WT cells (Fig. 3 E), which is consistent with NFAT1 activation downstream of Ca2+ up-regulation (Rao and Hogan, 2009). Downstream of DAG, α-CD3/CD28-induced phosphorylation of PKCθ and IκB-α was reduced in PLCβ4-deficient CD8+ T cells compared with that in WT cells (Fig. 3, F and G). Moreover, phosphorylation of MAPKs, such as ERK, p38, and JNK, was also markedly decreased in PLCβ4-deficient CD8+ T cells (Fig. 3 H). On the other hand, PLCβ4-deficient CD4+ T cells showed intact phosphorylation of SLP76, Lck, and PLCγ1 as well as PKCθ, ERK, and JNK in response to α-CD3/CD28 stimulation (Fig. 3, I and J). Furthermore, PMA/ionophore-mediated phosphorylation of PKCθ, IκBα, JNK, and ERK was intact in PLCβ4-deficient CD8+ T cells (Fig. 3, K–M). Taken together, these data suggest that PLCβ4 is required for full activation of TCR signaling pathways downstream of IP3 generation in CD8+ T cells but not in CD4+ T cells.
Figure 3.
PLCβ4-deficient CD8+ T cells display impaired activation of signaling cascades downstream of IP3. (A) Purified CD8+ T cells from WT or PLCβ4-deficient mice were stimulated with anti-CD3 (5 µg) and anti-CD28 (2 µg/ml) for indicated times. Indicated proteins were detected by Western blotting using specific phospho- or nonphospho Abs. (B) Concentrations of PIP2 in the lysates of purified CD8+ T cells from WT and PLCβ4-deficient mice were measured by ELISA. (C) Purified CD8+ T cells from WT and PLCβ4-deficient mice were stimulated with indicated doses of anti-CD3 and anti-CD28 for 1 h and lysed. IP1 levels were determined by ELISA. (D) Purified CD8+ T cells from WT or PLCβ4-deficient mice were loaded with Fluo-8 AM and stimulated with anti-CD3 (1 µg), followed by cross-linking (arrowhead). Ca2+ flux was measured by flow cytometry. The x axis shows real-time Ca2+ release followed for 300 s, and the y axis shows the intensity of the increase in intracellular Ca2+ concentration. MFI, mean fluorescence intensity. (E–H) Purified CD8+ T cells from WT or PLCβ4-deficient mice were stimulated with anti-CD3 (5 µg) and anti-CD28 (2 µg/ml) for indicated times. (E) Dephosphorylation of NFAT1 was detected by Western blotting with ant-NFAT1. (F and G) The phosphorylation of PKCθ (F) and IκBα (G) was determined by Western blotting with indicated Abs. (H) Activation of ERK, p38, and JNK was detected by Western blotting using specific phospho- or nonphospho Abs. (I and J) Purified CD4+ T cells from WT or PLCβ4-deficient mice were stimulated with anti-CD3 (5 µg) and anti-CD28 (2 µg/ml) for various times. (I) The phosphorylation of PKCθ was determined by Western blotting with indicated Abs. (J) Activation of ERK and p38 was detected by Western blotting using specific phospho Abs. (K–M) Purified CD8+ T cells from WT or PLCβ4-deficient mice were stimulated with PMA (50 ng/ml) and ionophore (1 µg/ml) for indicated times. The phosphorylation of PKCθ (K), IκBα (L), and MAP kinases such as ERK, p38, and JNK (M) were detected by Western blotting using specific phospho Abs. Indicated values are means ± SD of three biological replicates (B and C). *, P < 0.05; ***, P < 0.001, N.S., nonsignificant. Data are representative of three (A and D–H) or two (I–M) independent experiments.
PLCβ4-mediated activation of CD8+ T cells depends on interaction with the cytoplasmic tail of CD8α
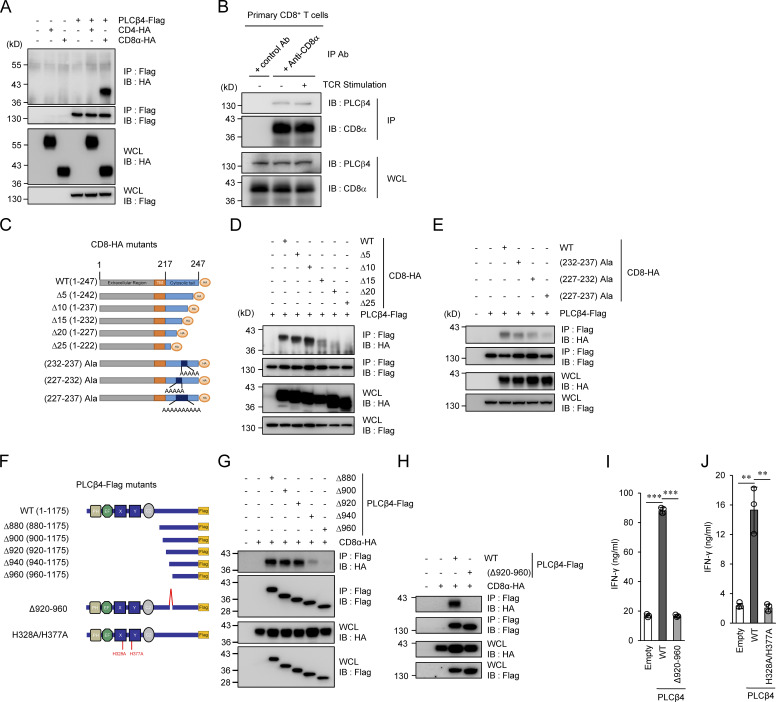
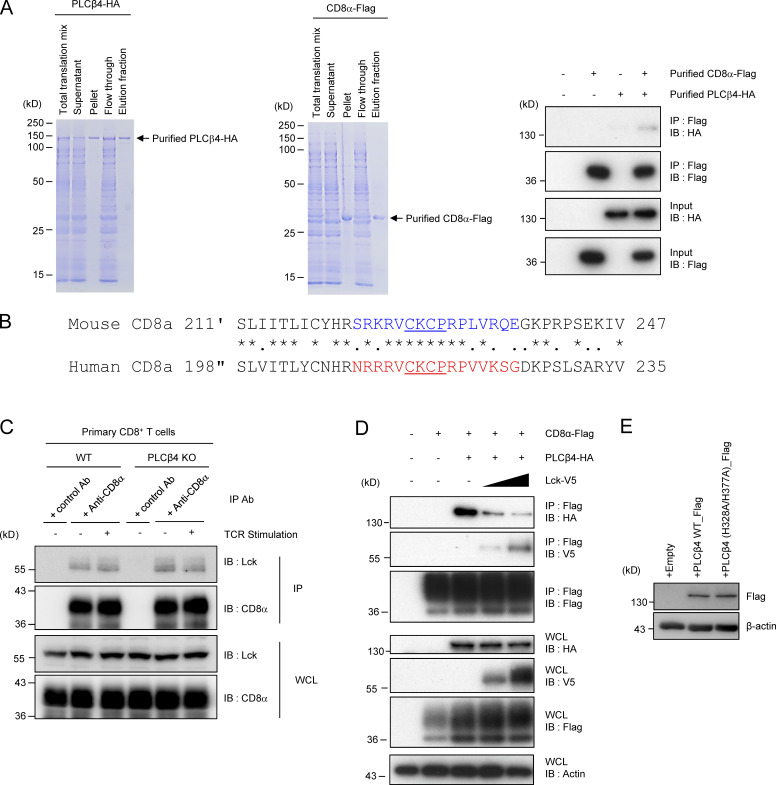
Next, we explored the molecular mechanism by which PLCβ4 is specifically involved in CD8+ T cells but not in CD4+ T cells. Our data suggested that PLCβ4 deficiency affects TCR signaling upstream of DAG (Fig. 3, A–D). However, the signaling molecules except for CD4 and CD8 are shared in CD4+ T cells and CD8+ T cells (Gaud et al., 2018). Therefore, we first started to examine the PLCβ4 interactions with CD4 or CD8α. Flag-tagged PLCβ4 coprecipitated with HA-tagged CD8α, but not with HA-tagged CD4 (Fig. 4 A). Furthermore, regardless of the TCR stimulation, endogenous CD8α coprecipitated with endogenous PLCβ4 in primary CD8+ T cells (Fig. 4 B). In addition, recombinant purified Flag-tagged CD8α proteins also coprecipitated with purified HA-tagged PLCβ4 proteins (Fig. S4 A). To identify which region of CD8α is responsible for the interaction with PLCβ4, we constructed several CD8α-deletion mutants lacking the cytoplasmic C-terminus and examined their interactions with PLCβ4 (Fig. 4 C). HA-tagged CD8α Δ15, Δ20, and Δ25 mutants showed impaired, greatly impaired, and nonexistent interactions, respectively, with Flag-tagged PLCβ4 (Fig. 4 D). Furthermore, when we performed alanine scanning around the region, the interaction between the section of CD8α containing alanines (aa 227–237) with PLCβ4 was dramatically reduced (Fig. 4 E), suggesting that the C-terminal region between the 227- and 237-aa positions may be important for interaction with PLCβ4. Amino acid sequences of the cytoplasmic C-terminus tails of mouse and human CD8α are well conserved. In particular, the PLCβ4 binding regions displays 71% similarity (Fig. S4 B). Since the PLCβ4 binding motif overlaps with the zinc clasp structure that tethers LCK to CD8α (Fig. S4 B; Kim et al., 2003), we next examined whether Lck and PLCβ4 compete to associate with CD8α (Fig. S4, C and D). In primary CD8+ T cells, association of CD8α with Lck was unaffected in the presence or absence of PLCβ4 in primary CD8+ T cells (Fig. S4 C). On the other hand, when we expressed V5-tagged Lck in a dose-dependent manner together with Flag-tagged CD8α and HA-tagged PLCβ4, the amounts of the precipitated PLCβ4 with CD8α were gradually reduced (Fig. S4 D), suggesting that the competition between PLCβ4 and Lck in their interaction with CD8α can happen.
Figure 4.
PLCβ4 binds to cytoplasmic tail of CD8α but not CD4. (A) Lysates of 293T cells transiently cotransfected with the indicated expression vectors were immunoprecipitated (IP) with anti-Flag and detected by Western blot (IB) with the indicated Abs. (B) Lysates of WT primary CD8+ T cells unstimulated or stimulated with anti-CD3 (5 µg/ml)/anti-CD28 (2 µg/ml) for 30 min were immunoprecipitated with the anti-CD8α or control IgG and detected by Western blot with the indicated Abs. (C) Schematic representation of various mutants of CD8α. (D and E) Lysates of 293T cells transiently cotransfected with Flag-tagged PLCβ4 and HA-tagged indicated mutants of CD8α vectors were immunoprecipitated with the anti-Flag and detected by Western blot with the indicated Abs. (F) Schematic representation of various mutants of PLCβ4. (G and H) Lysates of 293T cells transiently cotransfected with HA-tagged CD8α and Flag-tagged indicated deletion of PLCβ4 vectors were immunoprecipitated with the anti-Flag and detected by Western blot with the indicated Abs. (I and J) Purified CD8+ T cells from PLCβ4-deficient mice were retrovirally transduced with empty, PLCβ4 (WT), PLCβ4 (Δ920–940), or a catalytically inactive mutant of PLCβ4 (H328A/H377A) construct. The transduced cells were stimulated with anti-CD3 (0.5 µg) and anti-CD28 (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. Indicated values are means ± SD of three biological replicates (I and J). **, P < 0.01; ***, P < 0.001. Data are representative three independent experiments (A, B, D, E, G, and H).
Figure S4.
Assessment of possible competition between PLCβ4 and Lck in their interaction with CD8α. (A) Coomassie blue staining of the purified His-tagged recombinant PLCβ4-HA (left) and CD8α-Flag protein. The PLCβ4-HA (100 ng) and/or CD8α-Flag (100 ng) proteins were immunoprecipitated (IP) with anti-Flag and detected by Western blot (IB) with the indicated Abs. (B) Comparison of amino acid sequences for cytoplasmic tails of mouse and human CD8α. Colored or underlined amino acids denote the PLCβ4 binding region or the zinc clasp structure, respectively. (C) Lysates of WT or PLCβ4-deficient primary CD8+ T cells unstimulated or stimulated with anti-CD3 (5 µg/ml)/anti-CD28 (2 µg/ml) for 30 min were immunoprecipitated with anti-CD8α and detected by Western blot with the indicated Abs. (D) Lysates of 293T cells transiently cotransfected with the indicated expression vectors were immunoprecipitated with anti-Flag and detected by Western blot with the indicated Abs. WCL, whole cell lysates. (E) Purified CD8+ T cells from PLCβ4-deficient mice were transduced with empty, PLCβ4 (WT), or a catalytically inactive mutant of PLCβ4 (H328A/H377A) construct. Protein levels of PLCβ4 (WT) and mutant of PLCβ4 (H328A/H377A) in the indicated lysates were determined by Western blotting with indicated Abs. Data are representative of two (E) or three (A, C, and D) independent experiments.
We next explored the region of PLCβ4 required for CD8α binding. All PLCs including PLCβ4 share a highly conserved catalytic core containing an N-terminal pleckstrin homology domain, an EF-hands motif, a split X+Y catalytic domain, and a C2 domain (Kadamur and Ross, 2013). PLCβ isozymes have an additional C-terminal extension (Lyon and Tesmer, 2013; Suh et al., 2008). We constructed a series of PLCβ4-deletion mutants lacking the N-terminus and then performed immunoprecipitation (Fig. 4 F). Although Flag-tagged PLCβ4 mutants lacking aa 1–880, 1–900, and 1–920 (named Δ880, Δ900, and Δ920 mutants, respectively) associated with HA-tagged CD8α, PLCβ4 mutants lacking aa 1–940 and 1–960 (named Δ940 and Δ960) severely or completely lacked this interaction (Fig. 4, G and H). Moreover, the PLCβ4 deletion mutant lacking the internal 40 amino acids between aa 920 and 960 (named Δ920–960) resulted in loss of CD8α binding, indicating that aa 920–960 in the C-terminal extension of PLCβ4 are critical to CD8α binding (Fig. 4, G and H). Indeed, ectopic expression of PLCβ4 Δ920–960 in PLCβ4-deficient CD8+ T cells failed to reconstitute IFN-γ production (Fig. 4 I). Collectively, these results indicate that interaction of the C-terminal region of PLCβ4 with the cytoplasmic region of CD8α is responsible for the specific participation of PLCβ4 in activation of CD8+ T cells.
PLCβ4 lipase activity is essential for CD8+ T cell activation
Next, whether PLCβ4 lipase activity is required for CD8+ T cell activation was addressed (Fig. 4, F and J; and Fig. S4 E). PLCβ4-deficient CD8+ T cells were transduced with an empty vector, WT PLCβ4, or a catalytically inactive PLCβ4 mutant in which His328 and His377 were replaced with Ala, because both PLCβ1 and PLCδ1 mutants possessing this replacement in the corresponding His residues have been shown to be catalytically inactive (Ramazzotti et al., 2008). Reintroduction of WT PLCβ4 into PLCβ4-deficient CD8+ T cells up-regulated IFN-γ production. In contrast, introduction of the H328A/H377A mutants of PLCβ4 did not promote IFN-γ production despite expression in PLCβ4-deficient CD8+ T cells (Fig. 4, F and J), whereas the protein expression levels of the H328A/H377A mutants of PLCβ4 were comparable to those of WT PLCβ4 (Fig. S4 E), indicating that PLCβ4 lipase activity is required for activation of CD8+ T cells.
The GNAQ-PLCβ4 axis is important for TCR-mediated activation of CD8+ T cells
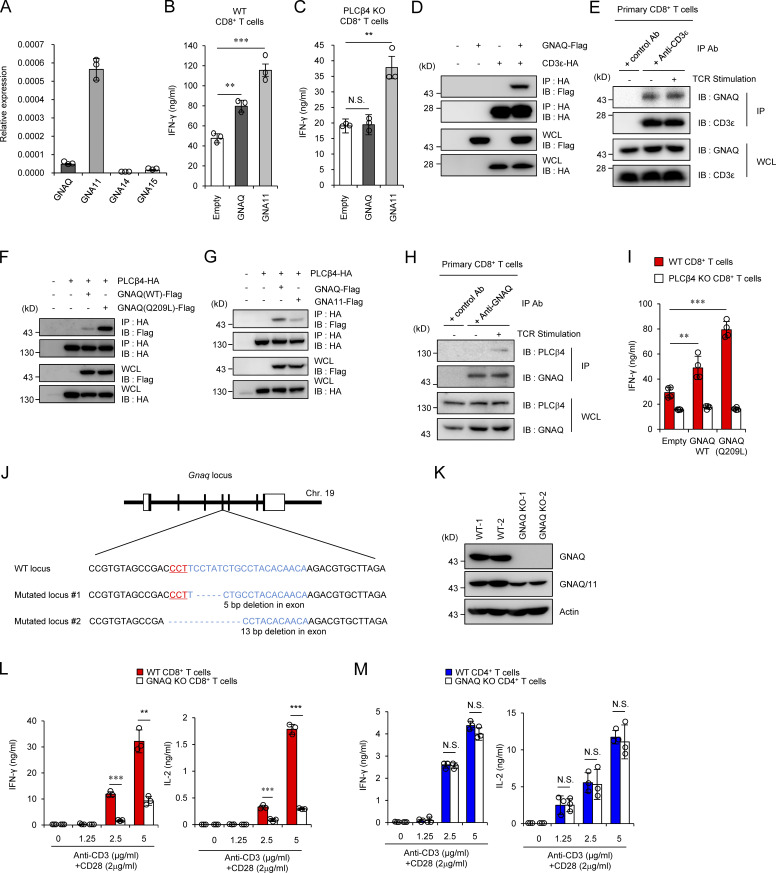
Next, we explored the mechanism by which PLCβ4 is activated in CD8+ T cells. Since the PLCβ isozymes have been shown to be activated by the heterotrimeric G protein α-subunit (GNA) family consisting of GNAQ, GNA11, GNA14, and GNA15 (Smrcka and Sternweis, 1993), we checked the mRNA expression levels of the GNA proteins in CD8+ T cells by quantitative PCR. Among the proteins, GNAQ and GNA11 were mainly expressed in CD8+ T cells (Fig. 5 A). To test whether GNAQ and GNA11 affect activation of PLCβ4 in CD8+ T cells, we ectopically expressed GNAQ or GNA11 in WT and PLCβ4-deficient CD8+ T cells by retroviral transduction (Fig. 5, B and C). Although the ectopic expression of GNAQ in WT cells significantly up-regulated IFN-γ production, overexpression of GNAQ in PLCβ4-deficient CD8+ T cells failed to do so (Fig. 5, B and C). In contrast, introduction of GNA11 promoted IFN-γ production in both WT and PLCβ4-deficient CD8+ T cells (Fig. 5, B and C), suggesting that GNAQ but not GNA11 might be involved in PLCβ4-dependent CD8+ T cell activation. In addition, we confirmed that GNAQ interacts with CD3ε (Fig. 5 D), as previously reported (Stanners et al., 1995). Moreover, regardless of the TCR stimulation, endogenous CD3ε bound to endogenous GNAQ in primary CD8+ T cells (Fig. 5 E). However, GNAQ did not interact with CD4 or CD8α (data not shown). Next, interactions between PLCβ4 and GNAQ or GNA11 were examined by coimmunoprecipitation assays. HA-tagged PLCβ4 associated with Flag-tagged GNAQ (Fig. 5 F). Moreover, PLCβ4 preferentially coprecipitated with GNAQ rather than GNA11 (Fig. 5 G). Moreover, a constitutively active mutant of GNAQ (Q209L GNAQ) exhibited remarkably enhanced interaction with PLCβ4 compared with that of WT GNAQ (Fig. 5 F). In addition, endogenous GNAQ associated with endogenous PLCβ4 in primary CD8+ T cells in a manner dependent on TCR stimulation (Fig. 5 H). Furthermore, introduction of Q209L GNAQ to WT CD8+ T cells further enhanced IFN-γ production in response to α-CD3 compared with introduction of WT GNAQ (Fig. 5 I). On the other hand, the enhancing effect of Q209L GNAQ was not observed in PLCβ4-deficient CD8+ T cells (Fig. 5 I), suggesting that GNAQ is located upstream of PLCβ4. To formally address the involvement of GNAQ in CD8+ T cell activation, we generated GNAQ-deficient mice by CRISPR/Cas9-mediated genome editing (Fig. 5, J and K). Similar to those in PLCβ4-deficient mice, CD4+ or CD8+ T cells in GNAQ-deficient mice showed no abnormality in composition of CD44 and CD62L (Fig. S3, A and B). We then assessed the cytokine production of GNAQ-deficient CD4+ and CD8+ T cells in response to α-CD3 (Fig. 5, L and M). Reminiscent of the phenotypes of PLCβ4-deficient CD8+ T cells, IFN-γ and IL-2 production were dramatically decreased in GNAQ-deficient CD8+ T cells (Fig. 5 L), while production in GNAQ-deficient CD4+ T cells was intact (Fig. 5 M). Taken together, these data strongly indicate that the GNAQ-PLCβ4 axis is involved in the activation of CD8+ T cells.
Figure 5.
The GNAQ-PLCβ4 axis is required for CD8+ T cell activation. (A) Quantitative PCR analysis of the expression of indicated GNA mRNA in WT CD8+ T cells. (B and C) Purified CD8+ T cells from WT (B) and PLCβ4-deficient (C) mice were transduced with empty, GNAQ, and GNA11 construct. The cells were stimulated with anti-CD3 (0.5 µg) and anti-CD28 (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. (D, F, and G) Lysates of 293T cells transiently cotransfected with the indicated expression vectors were immunoprecipitated (IP) with anti-HA and detected by Western blot (IB) with the indicated Abs. (E and H) Lysates of WT primary CD8+ T cells unstimulated or stimulated with anti-CD3 (5 µg/ml)/anti-CD28 (2 µg/ml) for 30 min were immunoprecipitated with anti-CD3ε or control IgG (E), and with anti-GNAQ or control IgG (H). The immunoprecipitated samples were detected by Western blot with the indicated Abs. (I) Purified CD8+ T cells from WT or PLCβ4-deficient mice were retrovirally transduced with empty, GNAQ (WT), or the constitutively active mutant of GNAQ (Q209L). The transduced cells were stimulated with anti-CD3 (0.5 µg/ml) and anti-CD28 (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. (J) The gene-targeting strategy for Gnaq locus by Cas9-mediated genome editing. Target sequences for gRNA were designed in the fourth exons of the Gnaq gene. Chr, chromosome. (K) WT and GNAQ-deficient tail fibroblasts were lysed, and the lysates were detected by Western blotting with the indicated Abs. (L and M) Purified CD8+ T cells (L) and CD4+ T cells (M) from WT and GNAQ-deficient mice were stimulated with indicated doses of anti-CD3 and anti-CD28 for 72 h. Concentrations of IFN-γ and IL-2 in the culture supernatants were measured by ELISA. Indicated values are means ± SD of three biological replicates (A–C, I, L, and M). **, P < 0.01; ***, P < 0.001; N.S., nonsignificant. Data are representative two (K) or three (D–H) independent experiments.
Phosphorylation of T886 and S890 positions on PLCβ4 downstream of GNAQ is required for CD8+ T cell activation
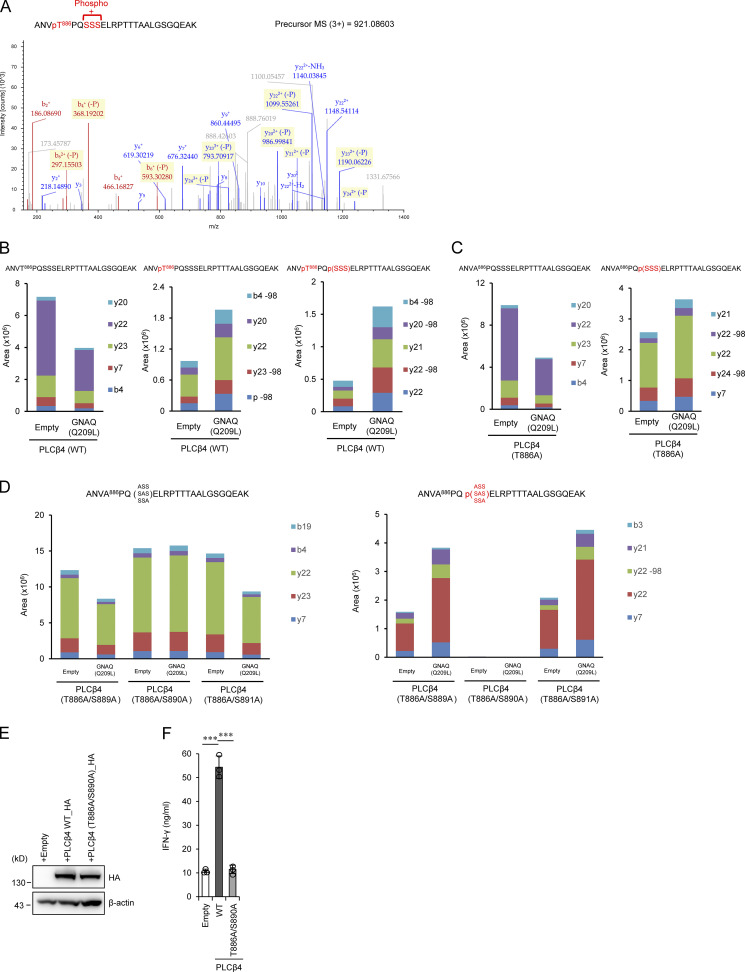
We next explored potential posttranslational modifications on PLCβ4 in 293T cells overexpressing the constitutively active GNAQ (GNAQ Q209L) mutant by mass spectrometry (MS) analysis. Overexpression of the GNAQ Q209L induced a single phosphorylation at threonine 886 (T886) or double phosphorylation at T886 and a serine in the serine 889 (S889), 890 (S890), or 891 (S891) sites on PLCβ4 (Fig. 6, A and B). T886A mutation resulted in a single phosphorylation at S889, S890, or S891, indicating that T886 is phosphorylated (Fig. 6 C). To further elucidate which serine of S889, S890, or S891 is phosphorylated in addition to T886, we generated T886A/S889A, T886A/S890A, and T886A/S891A PLCβ4 mutants and tested the phosphorylation by the GNAQ Q209L overexpression. We found that, although the T886A/S889A and T886A/S891A PLCβ4 mutants are still phosphorylated, the T886A/S890A mutant is not phosphorylated at all in response to the GNAQ Q209L overexpression (Fig. 6 D), indicating that T886 and S890 positions on PLCβ4 are phosphorylated downstream of GNAQ. Whether phosphorylation of T886/S890 sites on PLCβ4 is important for CD8+ T cell activation was further assessed by IFN-γ production from PLCβ4-deficient CD8+ T cells reconstituted with WT PLCβ4 or the T886A/S890A mutant (Fig. 6, E and F). Although reintroduction of WT PLCβ4 and the T886/S890A mutant in PLCβ4-deficient CD8+ T cells displayed comparable protein expression (Fig. 6 E), the T886A/S890A-reconstituted PLCβ4-deficient CD8+ T cells could not produce IFN-γ comparably to WT PLCβ4-reconstituted cells (Fig. 6 F), indicating that phosphorylation of T886 and S890 positions on PLCβ4 downstream of GNAQ is important for CD8+ T cell activation.
Figure 6.
GNAQ-induced PLCβ4 phosphorylation is important for CD8+ T cell activation. (A) 293T cells transiently cotransfected with HA-tagged WT PLCβ4 and Flag-tagged GNAQ (Q209L) were subjected to immunoprecipitation with anti-HA antibody followed by tryptic digestion and LC-MS/MS analysis. The MS/MS spectrum suggested that phosphorylation of PLCβ4 occurs at T886 and S889, S890, or S891. (B–D) 293T cells transiently cotransfected with HA-tagged WT (B), T886A (C), or T886A/S889A, T886A/S890A, and T886A/S891A PLCβ4 (D) and Flag-tagged GNAQ (Q209L) or the empty vector were subjected to immunoprecipitation with anti-HA antibody followed by tryptic digestion. Phosphopeptides and nonphosphopeptides of the A883-K908 peptides were quantified by targeted MS using the parallel reaction monitoring method. It should be noted that the phosphopeptide was completely eliminated by T886A/S890A mutation. (E) Purified CD8+ T cells from PLCβ4-deficient mice were transduced with empty, PLCβ4 (WT), or PLCβ4 (T886A/S890A) construct. Protein levels of PLCβ4 (WT) and mutant of PLCβ4 (T886A/S890A) in the indicated lysates were determined by Western blotting with indicated Abs. (F) Purified CD8+ T cells from PLCβ4-deficient mice were retrovirally transduced with empty, PLCβ4 (WT), or mutant of PLCβ4 (T886A/S890A) construct. The transduced cells were stimulated with anti-CD3 (0.5 µg) and anti-CD28 (1 µg/ml) for 72 h. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. Indicated values are means ± SD of three biological replicates (F). ***, P < 0.001. Data are representative of two independent experiments (A–E).
In vitro and in vivo OVA peptide-mediated CD8+ T cell activation requires PLCβ4
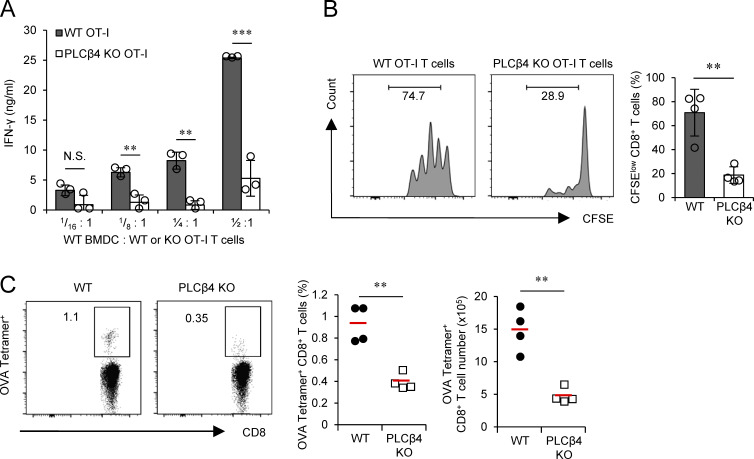
Next, we examined whether CD8+ T cell activation by MHC with antigenic peptide requires PLCβ4 expression in CD8+ T cells. PLCβ4-deficient mice possessing OVA-specific OT-I TCRs were generated. CD8+ T cells were isolated and cocultured with OVA peptide–pulsed WT dendritic cells and tested for IFN-γ production and proliferation of CD8+ T cells (Fig. 7 A). IFN-γ production from PLCβ4-deficient OT-I cells cocultured with WT dendritic cells was significantly reduced compared with that in WT OT-I cells (Fig. 7 A). Next, we transferred WT or PLCβ4-deficient OT-I cells into WT mice to compare their proliferation in response to immunization of OVA peptide in vivo (Fig. 7 B). We found that PLCβ4-deficient OT-I cells proliferated less than did WT cells in the mice immunized with OVA peptide (Fig. 7 B), indicating that PLCβ4 is physiologically required for CD8+ T cell activation via OVA pMHCI. We then examined whether the CD8+ T cell–dependent adaptive immune response is impaired in PLCβ4-deficient mice. 14 d after immunization with OVA peptide, the frequencies of OVA-specific CD8+ T cells in the spleens of WT and PLCβ4-deficient mice were compared (Fig. 7 C). Spleens from PLCβ4-deficient mice contained significantly decreased percentages and numbers of OVA-specific CD8+ T cells than did those from WT mice (Fig. 7 C). Taken together, these data suggest that PLCβ4 is required for activation of CD8+ T cells via physiological antigen presentation.
Figure 7.
PLCβ4 is important for CD8+ T cell–mediated acquired immunity to OVA peptide. (A) Purified WT or PLCβ4-deficient OT-I CD8+ T cells were cocultured with WT BMDCs for 3 d at indicated ratios. Concentrations of IFN-γ in the culture supernatants were measured by ELISA. (B) CFSE-labeled WT or PLCβ4-deficient OT-I CD8+ T cells were adoptively transferred into WT mice, which were then injected subcutaneously with OVA (50 µg). After 2 d, the fluorescence intensity of CFSE-labeled OT-I CD8+ T cells in the spleen was analyzed by flow cytometry. (C) WT and PLCβ4-deficient mice were injected subcutaneously with OVA (50 µg) plus cGAMP (10 µg) on days 1 and 10. 10 d after the second immunization, the percentages and numbers of OVA-specific CD8+ T cells in the spleen from WT or PLCβ4-deficient mice were measured by flow cytometry. Indicated values are means ± SD of three biological replicates (A). **, P < 0.01; ***, P < 0.001; N.S., nonsignificant. Data are cumulative of four independent experiments (B and C).
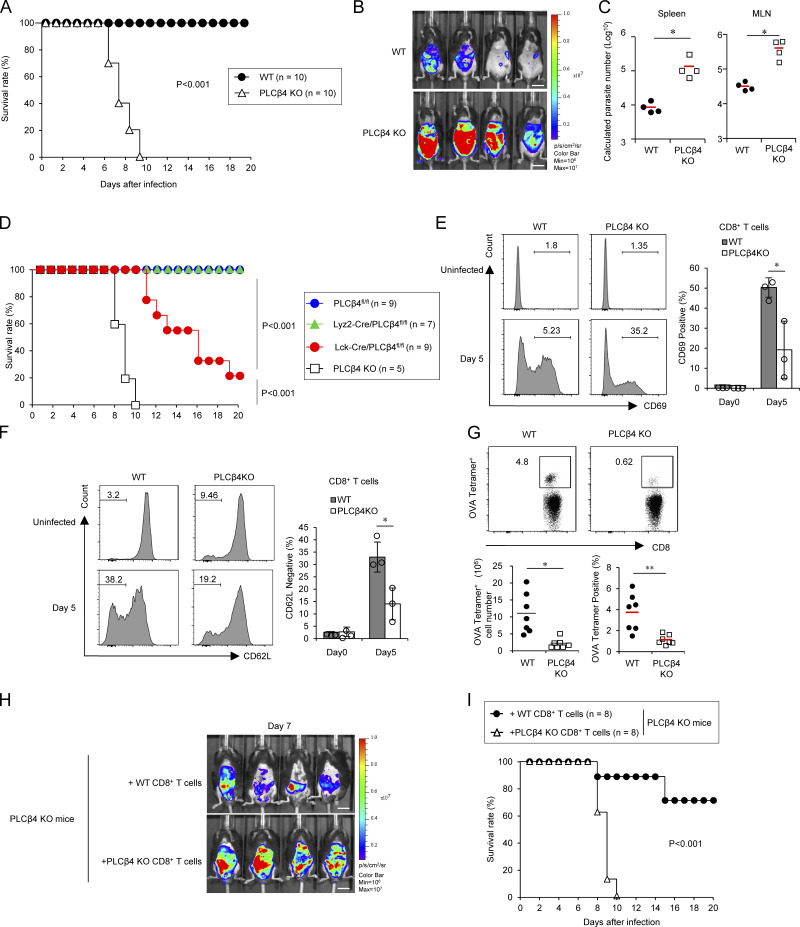
Defective CD8+ T cell activation leads to high susceptibility to Toxoplasma gondii in PLCβ4-deficient mice
To investigate the role of PLCβ4 in host defense, we challenged PLCβ4-deficient and WT mice with luciferase-expressing T. gondii, an important human and animal protozoan pathogen that causes lethal toxoplasmosis in immunocompromised individuals, such as those with AIDS or organ/bone marrow transplants (Boothroyd, 2009; Sasai et al., 2018). We found that PLCβ4-deficient mice were highly susceptible to T. gondii infection at acute phase when compared with WT mice. While WT mice survived, PLCβ4-deficient mice succumbed by day 10 (Fig. 8 A). Parasite burdens were significantly higher in PLCβ4-deficient mice than in WT mice (Fig. 8 B). Moreover, spleens and mesenteric lymph nodes from PLCβ4-deficient mice contained higher parasite numbers than did those from WT mice (Fig. 8 C). Despite the high mortality of T. gondii–infected PLCβ4-deficient mice, whether this high susceptibility was due to PLCβ4 deficiency in CD8+ T cells was unclear. Thus, we examined whether PLCβ4 depletion in T cells leads to loss of resistance to T. gondii infection (Figs. 8 D and S5 A). Lck-Cre/PLCβ4fl/fl mice were more susceptible to T. gondii infection than were control PLCβ4fl/fl mice (Fig. 8 D). In contrast, myeloid-specific PLCβ4 depletion did not affect host resistance to T. gondii (Figs. 8 D and S5 B), indicating that PLCβ4 expression in T cells but not in the myeloid lineage is important to the antiparasitic response. Next, we examined CD8+ T cell responses during T. gondii infection in WT and PLCβ4-deficient mice (Fig. 8, E and F). Regarding activation of CD8+ T cells, we found that the percentages of CD69+ and CD62Llow CD8+ T cells in the spleens of PLCβ4-deficient mice 5 d after T. gondii infection were significantly lower than those in the spleens of WT mice (Fig. 8, E and F). To analyze whether PLCβ4 deficiency affects the T. gondii–specific CD8+ T cell–dependent immune response, WT and PLCβ4-deficient mice were infected with irradiated T. gondii expressing OVA. We then determined the frequency and number of OVA-specific CD8+ T cells in the spleens by OVA-tetramer staining 14 d after infection (Fig. 8 G). The spleens of OVA-expressing T. gondii–infected PLCβ4-deficient mice contained significantly lower percentages and numbers of OVA-tetramer–positive CD8+ T cells than did those of WT mice (Fig. 8 G). On the other hand, activation and percentages or numbers of OVA-specific CD4+ T cells in the spleens after T. gondii infection were comparable between WT and PLCβ4-deficient mice (Fig. S5, C and D). To examine whether increased mortality in PLCβ4-deficient mice following T. gondii infection was caused by insufficient CD8+ T cell responses, we adoptively transferred purified polyclonal WT or PLCβ4-deficient CD8+ T cells into PLCβ4-deficient mice before infection with T. gondii and then monitored survival rates and in vivo parasite burdens (Fig. 8, H and I). In vivo parasite burdens were lower in mice that received WT CD8+ T cells than in those receiving PLCβ4-deficient CD8+ T cells (Fig. 8 H). Furthermore, PLCβ4-deficient mice receiving WT CD8+ T cells showed significantly prolonged survival compared with mice receiving PLCβ4-deficient CD8+ T cells (Fig. 8 I). Collectively, PLCβ4 deficiency leads to defective activation of CD8+ T cells and loss of host defense.
Figure 8.
PLCβ4 is required for CD8+ T cell–mediated host defense against T. gondii. (A) WT or PLCβ4-deficient mice (n = 10 per group) were infected with 1 × 105 T. gondii, and the survival rates were monitored for 20 d. P < 0.001 (WT vs. PLCβ4-deficient, log-rank test). (B) WT or PLCβ4-deficient mice (n = 4 per group) were infected with 1 × 105 Pru T. gondii expressing luciferase, and the progress of infection was assessed by bioluminescence imaging on day 7 after infection. The color bar indicates photon emission during a 60-s exposure. Scale bar: 1 cm. p/cm2/s/sr: photon per square centimeter per second per steradian. (C) Quantification of parasites in indicated tissues from mice (n = 4 per group) on day 8 after infection using the standard curve (in Fig. S5 E). *, P < 0.05. (D) Survival analysis of PLCβ4fl/fl (n = 9), Lck-Cre/PLCβ4fl/fl (n = 9), Lyz2-Cre/PLCβ4fl/fl (n = 7), or PLCβ4-deficient (n = 5) mice after T. gondii infection. P < 0.001 (PLCβ4fl/fl vs. Lck-Cre/PLCβ4fl/fl, log-rank test); P < 0.001 (PLCβ4 KO vs. Lck-Cre/PLCβ4fl/fl, log-rank test). (E and F) WT or PLCβ4-deficient mice (n = 3 per group) were infected with 1 × 105 T. gondii. After 5 d, the percentages of CD69+ (E) and CD62Llow (F) population on CD8+ T cells in the spleens from parasite-infected WT or PLCβ4-deficient mice were measured by flow cytometry. (G) WT or PLCβ4-deficient mice (n = 7 per group) were infected with 1 × 105 OVA expressing irradiated T. gondii. 14 d after infection, the percentages and cell numbers of OVA tetramer-positive CD8+ T cells in the spleens from parasite-infected WT or PLCβ4-deficient mice were measured by flow cytometry. (H) Purified WT or PLCβ4-deficient CD8+ T cells were adoptively transferred into PLCβ4-deficient mice (n = 4 per group). Mice were infected with luciferase-expressing T. gondii, and the progress of the infection was assessed by bioluminescence imaging on day 7 after infection. Color scales indicate photon emission during a 60-s exposure. Scale bar: 1 cm. (I) Purified WT or PLCβ4-deficient CD8+ T cells were adoptively transferred into PLCβ4-deficient mice (n = 8 per group). Mice were infected with T. gondii, and the survival rates were monitored for 20 d. P < 0.001 (mice receiving WT CD8+ T cells vs. mice receiving PLCβ4-deficient CD8+ T cells, log-rank test). Indicated values are means ± SD of three biological replicates (E and F). *, P < 0.05; **, P < 0.01; N.S., nonsignificant. Data are cumulative (A, D, G, and I) or representative (B, C, and H) of three independent experiments.
Figure S5.
Generation of T cell– or myeloid-specific PLCβ4-deficient mice. (A and B) BMDM, CD4+, or CD8+ T cells from WT, PLC4β4fl/fl, Lyz2-Cre/PLCβ4fl/fl, or Lck-Cre/PLCβ4fl/fl mice were lysed. PLCβ4 protein levels in the indicated lysates were determined by Western blotting with indicated Abs. (C) WT or PLCβ4-deficient mice (n = 3 per group) were infected with 1 × 105 T. gondii. After 5 d, the percentages of CD69+ (left) and CD62Llow (right) population on CD4+ T cells in the spleens from parasites infected WT or PLCβ4-deficient mice were measured by flow cytometry. (D) WT or PLCβ4-deficient mice (n = 6 per group) were infected with 1 × 105 OVA expressing irradiated T. gondii. 14 d after infection, the percentages and cell numbers of OVA tetramer-positive CD4+ T cells in the spleens from parasite-infected WT or PLCβ4-deficient mice were measured by flow cytometry. (E) Luciferase activities of serial dilutions of Pru expressing luciferase (103–107) were plotted. A standard curve of luciferase activity (x axis) and parasite number (y axis) was generated by the data plots. Indicated values are means ± SD of three biological replicates (C). N.S., nonsignificant. Data are cumulative of three independent experiments (D) and representative of two (A, B) or three (E) independent experiments.
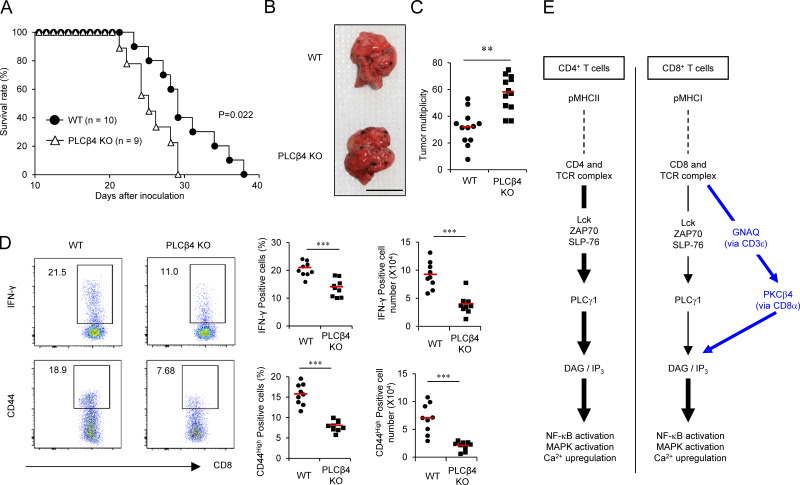
PLCβ4 is required for CD8+ T cell–mediated antitumor immunity
Since CD8+ T cells are essential for antitumor immunity (Golstein and Griffiths, 2018), we finally examined the importance of PLCβ4 to the antitumor immune response in a lung metastasis model using B16F10 mouse melanoma (Fig. 9, A–D). WT or PLCβ4-deficient mice were intravenously injected with B16F10 melanoma cells. We then monitored survival rate and lung metastasis. Notably, survival periods of tumor-injected PLCβ4-deficient mice were significantly shorter than those of tumor-injected WT mice (Fig. 9 A). Furthermore, PLCβ4-deficient mice showed significantly more lung metastasis than did WT mice (Fig. 9, B and C). Furthermore, the lungs of tumor-injected PLCβ4-deficient mice contained markedly lower percentages and numbers of IFN-γ–producing and CD44high CD8+ T cells than did those of WT mice (Fig. 9 D), indicating that the antitumor immune response required PLCβ4.
Figure 9.
PLCβ4 is involved in antitumor CD8+ T cell response. (A) B16F10 melanoma cells (2 × 105) were intravenously injected into WT (n = 10) or PLCβ4-deficient mice (n = 9), and the survival rates were monitored for 40 d. P = 0.022 (WT vs. PLCβ4-deficient, log-rank test). (B and C) B16F10 melanoma cells (2 × 105) were intravenously injected into WT or PLCβ4-deficient mice (n = 12 per group). After 21 d, representative images of lungs from WT or PLCβ4-deficient mice (B) and the numbers of metastatic nodules in the lungs from WT or PLCβ4-deficient mice (C) are shown. Scale bar: 1 cm. (D) B16F10 melanoma cells (2 × 105) were intravenously injected into WT or PLCβ4-deficient mice (n = 9 per group). After 14 d, the percentages of IFN-γ+ CD8+ T cells in the lungs from WT or PLCβ4-deficient mice were measured by intracellular staining. The percentages of CD44+ population on CD8+ T cells in the lungs from WT or PLCβ4-deficient mice were measured by flow cytometry. (E) Schematic of the GNAQ-PLCβ4 axis in CD8+ T cell TCR signaling. **, P < 0.01; ***, P < 0.001; N.S., nonsignificant. Data are cumulative (A, C, D) or representative (B) of three independent experiments.
Discussion
In the present study, we demonstrate that CD8+ T cells but not CD4+ T cells physiologically require PLCβ4 for the TCR signaling pathways downstream of IP3 and DAG to sufficiently mount host defense and antitumor adaptive immunity (Fig. 9 E). PLCγ1 is required for α-CD3/CD28–mediated production of IL-2 and IFN-γ and proliferation of both CD4+ and CD8+ T cells (Fu et al., 2010). Loss of PLCγ1 leads to profoundly defective Ca2+ up-regulation and activation of MAPKs and NF-κB (Fu et al., 2010). In contrast, PLCβ4 deficiency resulted in moderate defects in Ca2+ up-regulation, IP3 production, and activation of MAPKs and NF-κB. T cell–specific PLCγ1 deletion results in severe impairment of T cell development in the thymus (Fu et al., 2010). In addition, deficiency of any of Lck, ZAP70, SLP76, or LAT, which all are universally essential for the TCR signaling pathways in both CD4+ and CD8+ T cells, leads to defective T cell development in the thymus (Clements et al., 1998; Molina et al., 1992; Negishi et al., 1995; Zhang et al., 1999). On the other hand, the PLCβ4 mRNA and proteins in double-negative, double-positive, and CD8+ single-positive thymocytes were detected. In the present study, we could not reveal a defect in T cell development in the thymus of PLCβ4-deficient mice. To conclude that PLCβ4 deficiency does not affect T cell development, more extensive studies such as TCR repertoire analysis and phenotyping of different cellular compartments might be required in the future. The defects in PLCβ4-deficient CD8+ T cells in response to extremely strong CD3/CD28 stimulation were less obvious. Moreover, all of responses of PLCβ4-deficient CD8+ T cells were never abolished, albeit they were severely weakened, which contrasts with the previous finding of PLCγ1-deficient mice displaying stronger phenotypes, including defective thymic T cell development, than PLCβ4-deficient mice (Fu et al., 2010). Taken together, these findings suggest that the additional PLCβ4-dependent TCR signaling pathway may work in parallel with the basal PLCγ1-dependent pathway in CD8+ T cells under physiological strength of the stimulation, probably correlating the distinct consequence of thymic T cell development in PLCβ4-deficient or PLCγ1-deficient mice. Moreover, CD4+ T cells expressed markedly more PLCβ4 than CD8+ T cells. Although we could not identify the specific function of PLCβ4 in CD4+ T cells in the present study, further analysis might reveal a possible role of PLCβ4 in TCR signaling except for the response to α-CD3/CD28, or functions such as postselection TCR repertoire and effector responses in CD4+ T cells.
Although we detected interaction between purified PLCβ4 and CD8α proteins, the aa 920–960 in the C-terminal extension of PLCβ4 contain highly basic residues, leading to possible repulsion between the proteins. In fact, when we tested the interaction in 293T cells, we only tested the N-terminal deletion PLCβ4 mutants, since we could not express the C-terminal deletion mutants due to unknown reasons (data not shown). Since we were not able to examine the role of the very C-terminus of PLCβ4, we cannot exclude the possibility that unidentified C-terminal regions in addition to aa 920–960 of PLCβ4 are required for direct interaction with CD8α.
We found that the GNAQ-PLCβ4 axis is important for activation of CD8+ T cells. In addition to the mechanism dependent on protein tyrosine kinases, additional regulatory mechanisms involving heterotrimeric GTP-binding proteins in T cell activation have been reported (Anderson and Tsoukas, 1989; Cenciarelli et al., 1992; Graves and Cantrell, 1991; Imboden et al., 1986; O’Shea et al., 1987). TCR complexes have been shown to include a phosphoprotein containing a GTP-binding region of G proteins (Telfer and Rudd, 1991). Treatment with cholera toxin, which activates the stimulatory G protein of adenylate cyclase by catalyzing transfer of ADP-ribose to GNA (Gilman, 1984), inhibits α-CD3–induced proliferation and Ca2+ increase in T cells (Anderson and Tsoukas, 1989; Imboden et al., 1986). Moreover, treatment with GDPβS, an inhibitor of G proteins, abrogates anti-CD3–induced generation of inositol phosphates in peripheral T cells, whereas treatment with GTPγS and aluminum fluoride, which activates G proteins, induces phosphatidylinositol turnover, elevates cytoplasmic free calcium, and activates T cells (Graves and Cantrell, 1991; O’Shea et al., 1987).
With respect to roles of GNAs in T cell activation, Jurkat T cells expressing the dominant-negative form of GNA16 display impaired TCR signaling and cytokine production (Zhou et al., 1998). On the other hand, primary splenic T cells lacking GNAQ show reduced LAT phosphorylation and sustained ERK phosphorylation but increased production of cytokines, such as IL-2, IL-5, IL-12, and TNF-α (Ngai et al., 2008). In contrast, our GNAQ-deficient mice showed reduced IL-2 and IFN-γ production in CD8+ T cells but not in CD4+ T cells. This discrepancy might be due to different origins of the GNAQ-deficient mice or distinct protocols for T cell purification. We separately isolated CD4+ T cells and CD8+ T cells by flow cytometry and have not detected IL-12 production in these T cells. Furthermore, GNAQ and GNA11 are activated in response to α-CD3 stimulation and associate with the CD3ε subunit (Stanners et al., 1995). Moreover, bacterial superantigens activate T cells in a manner dependent on GNA11 (Bueno et al., 2006). Thus, accumulating evidence suggests involvement of G proteins in TCR signaling pathways; however, the physiological relevance of the GNA-PLCβ axis to TCR signaling pathways remained unclear. Herein, we found that GNAQ but not GNA11 activates CD8+ T cells in a manner dependent on PLCβ4. In addition, PLCβ4 preferentially associates with GNAQ rather than GNA11. Furthermore, we showed that GNAQ deficiency only affects activation of CD8+ T cells but not CD4+ T cells, which is reminiscent of PLCβ4 deficiency. Given the interaction of PLCβ4 with CD8α, PLCβ4 might be localized with GNAQ, which binds CD3ε, and activated by GNAQ in the CD3/TCR complex of CD8+ T cells. Moreover, PLCβ4-deficient OT-I CD8+ T cells exhibited impaired IFN-γ production and proliferation when cocultured with APCs expressing OVA pMHCI, suggesting that the GNAQ-PLCβ4 axis is physiologically operative in TCR activation of CD8+ T cells via pMHCI. In addition, a component of the heterometric G protein involving GNAQ is the β1γ2 subunit (Evanko et al., 2000; Grabocka and Wedegaertner, 2005). Given that PLCβ4 activation is shown to be dependent on GNAQ but independent of the βγ subunit (Kadamur and Ross, 2013; Lee et al., 1994), the βγ subunit may be not involved in the TCR activation of CD8+ T cells. Although overexpression of GNAQ but not GNA11 activates CD8+ T cells in a manner dependent on PLCβ4, GNA11 could associate with PLCβ4. Moreover, given that GNA11 is expressed more highly than GNAQ and is involved in bacterial superantigen-induced T cell activation (Bueno et al., 2006), GNA11 as well as GNAQ might possibly participate in TCR signaling pathway of CD8+ T cells under such a pathological condition.
We found that T886 and S890 sites are phosphorylated downstream of GNAQ, and that these phosphorylation sites are important for CD8+ T cell activation. It is of note that threonine/serine residues rather than tyrosine residues on PLCβ4 are targeted by GNAQ in TCR signaling of CD8+ T cells. The kinetics of T886/S890 phosphorylation in response to CD8+ T cell activation could be tested by generation of phospho-T886/S890 PLCβ4 specific antibody in the future. In addition, it is plausible to postulate that the serine/threonine kinase responsible for the T886/S890 phosphorylation on PLCβ4 exists and might be activated in response to CD3/CD28 stimulation in CD8+ T cells.
Herein, we demonstrated that PLCβ4 selectively interacts with CD8α but not with CD4, conferring the specificity of PLCβ4 in the TCR signaling pathways for CD8+ T cells. Since the proportion of cellular Lck binding to CD4 is 10–20-fold higher than that interacting with CD8α (Julius et al., 1993; Veillette et al., 1989), CD4/Lck may sufficiently activate TCR signaling pathways in CD4+ T cells, whereas CD8+ T cells might insufficiently transduce activation signals from the CD3/TCR complex only via CD8α/Lck and hence eventually require the additional signal via CD8α/PLCβ4 to boost the TCR signaling pathways. Furthermore, we found that PLCβ4 interacts with the cytoplasmic region of CD8α containing the CXCP motif, to which Lck also binds (Shaw et al., 1990; Turner et al., 1990). Although we did not detect increased binding of endogenous Lck to CD8α even in the absence of PLCβ4 in primary CD8+ T cells, increased Lck proteins reduced the PLCβ4 binding to CD8α in 293T cells. The discrepancy might be due to the different PLCβ4 expression levels in distinct cell types. Given that PLCβ4 binding motif overlaps with the zinc clasp structure that tethers Lck to CD8α (Shaw et al., 1990; Turner et al., 1990), competition between PLCβ4 and Lck in interaction with CD8α could happen, when PLCβ4 is abundantly present in the overexpression system in 293T cells. On the other hand, since the protein levels of Lck are much higher than those of PLCβ4 in primary CD8+ T cells, the lack of PLCβ4 might not be able to have an impact on the Lck interaction with CD8α. Furthermore, a previous study showed reciprocal regulation between tyrosine kinases and G proteins in human Jurkat T cells (Stanners et al., 1995). We found that PLCβ4 deficiency does not affect α-CD3–induced Lck phosphorylation, suggesting that the GNAQ-PLCβ4 axis does not regulate Lck activation in TCR signaling pathways in CD8+ T cells. It would be interesting to examine whether Lck regulates activation of the GNAQ-PLCβ4 axis in the future.
We showed that PLCβ4-deficient mice are highly susceptible to acute toxoplasmosis due to defective CD8+ T cell–mediated host defense. It has been shown that mice deficient in CD8+, but not CD4+, T cell functions are highly susceptible to acute toxoplasmosis (Casciotti et al., 2002; Combe et al., 2005; Lu et al., 2009), indicating that optimal PLCβ4-mediated CD8+ T cell activation is required for mounting anti–T. gondii acquired immunity at early stages of the infection. We demonstrated that PLCβ4-deficient mice are also susceptible to a mouse melanoma metastasis model. CD8+ T cells are also important for antitumor acquired immunity through the release of cytotoxic molecules such as granzymes and IFN-γ (Barnes and Amir, 2017; Fridman et al., 2012; Kaech and Cui, 2012). Given that CD8+ T cells play a critical role in inhibition of cancer metastasis (Lengagne et al., 2008), PLCβ4 deficiency in CD8+ T cells may lead to defective B16F10 melanoma-derived antigen-induced CD8+ T cell activation and hence reduced IFN-γ production.
In conclusion, we have identified PLCβ4 as a component in TCR signaling pathways for CD8+ T cells. Pharmacological activation of PLCβ4 may provide novel CD8+ T cell–specific therapeutics for infectious diseases and anticancer treatment.
Materials and methods
Cells, mice, and parasites
C57BL/6NCrSlc (C57BL/6N) mice were purchased from Japan SLC, Inc. All animal experiments were conducted with the approval of the Animal Research Committee of Research Institute for Microbial Diseases in Osaka University. Pru strains of T. gondii were maintained in Vero cells by biweekly passage in RPMI 1640 (Nacalai Tesque) supplemented with 2% heat-inactivated FBS (Gibco), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Nacalai Tesque). B16F10, Plate-E, and 293T cells were maintained in DMEM (Nacalai Tesque) supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Nacalai Tesque).
Reagents
For Western blot analysis, anti-PLCβ4 (sc-166131), anti-HA (sc-7392), anti-GNAQ (sc-136181), anti-ERK (sc-94), anti-p38 (sc-535), anti-JNK (sc-7345), anti-ZAP70 (sc-32760), and anti-PLCγ1 (sc-7290) were purchased from Santa Cruz Biotechnology. Anti-pZAP70 (#2717), anti-pSLP76 (#14745), anti-PKCθ (#9377), anti-pERK (#4370), anti–p-p38 (#4511), anti-pJNK (#4668), anti–pIκB-α (#9246), anti-NFAT1 (#5861), and anti-pPLCγ1 (#14008) were purchased from Cell Signaling Technology. Anti-FLAG (F1804), anti–β-actin (A1978), and anti–HA-agarose (A2095) were purchased from Sigma-Aldrich. Anti-V5 (PM003) was obtained from MBL. For T cell stimulation, anti-CD3ε (145-2C11; 100302) and anti-CD28 (37.51; 102112) were purchased from BioLegend. For flow cytometric analysis, anti-CD4 (12-0041-81), anti-CD8α (25-0081-81), anti-CD69 (17-0691-80), and anti–TNF-α (12-7321-81) were purchased from eBioscience. Anti-CD45 (103126), anti-CD25 (102022), and anti–IFN-γ (505816) were purchased from BioLegend. Anti-CD44 (561862), anti–IFN-γ (557735) and anti-CD62L (560513) were purchased from BD PharMingen. Recombinant mouse IFN-γ, IL-2, and IL-12 were obtained from Peprotech. PMA and calcium ionophore (A23187) were obtained from Sigma-Aldrich. Anti-Lck phosphor-Tyr394 (933102) was obtained from BioLegend. Anti-total Lck (ab3885) was obtained from Abcam. For immunoprecipitation of CD8α, anti-CD8α rat Ab (sc-18913) was obtained from Santa Cruz Biotechnology. For immunoblot of CD8α, anti-CD8α rabbit Ab (ab217344) was obtained from Abcam. Anti-GNAQ/11 antibody (13927-1-AP), which detects both GNAQ and GNA11, was obtained from Proteintech.
Generation of PLCβ4-deficient mice
The Plcb4 gene was isolated from genomic DNA extracted from embryonic stem (ES) cells (V6.5) by PCR using KOD FX NEO (Toyobo). The targeting vector was constructed by replacement of a 1.0-kb fragment of Plcb4 with a neomycin-resistance gene (neo) cassette driven by a phosphoglycerate kinase (PGK) promoter, which follows a splice acceptor (SA) sequence to trap the gene. In addition, a 1.0-kb fragment of Plcb4 genomic DNA containing exon 6 was inserted between two loxP sites in the targeting vector pKS-SApGKneo-DTA, which contains a diphtheria toxin A fragment driven by a PGK promoter. After the targeting vector was transfected into ES cells using electroporation, colonies resistant to G418 were selected and screened by PCR and Southern blotting as described previously (Yamamoto et al., 2011). Correctly targeted ES cells were injected into C57BL/6N blastocysts, and the chimeric male mice were tested for germline transmission by crossing them with female C57BL/6N mice to generate Plcb4gt/+ F1 mice. The heterozygous F1 mice were backcrossed eight times with the C57BL/6N strain. Plcb4gt/gt mice (PLCβ4-deficient mice) and their WT littermates from these intercrosses were used for experiments. To generate PLCβ4 conditional knockout mice, heterozygote Plcb4gt/+ mice were crossed with transgenic CAG-Flpo mice to remove the neo cassette franked by two FRT sequences to generate Plcb4flox/+ (PLCβ4fl/+ mice). PLCβ4fl/fl mice were crossed with Lck-Cre or Lyz2-Cre transgenic mice to generate T cell–specific or myeloid-specific PLCβ4-depleted mice, respectively.
Generation of GNAQ-deficient mice by CRISPR genome editing
T7 promoter was added to the guide RNA (gRNA) template using KOD FX NEO and primers GNAQ_T7gRNA_F and gRNA_common_R. The T7-transcribed GNAQ gRNA PCR products were gel-purified and used for the subsequent generation of gRNA. MEGAshortscript T7 (Life Technologies) was used for generation of the gRNA. mRNA encoding the RNA-guided DNA endonuclease Cas9 was generated by in vitro transcription using the mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies), and the template was amplified by PCR using pEF6-hCas9-Puro and the primers T7Cas9_IVT_F and Cas9_R and gel-purified. The synthesized gRNA and Cas9 mRNA were purified using the MEGAclear kit (Life Technologies) and eluted in RNase-free water (Nacalai Tesque). To obtain GNAQ-deficient mice, 6-wk-old female C57BL/6N mice were superovulated and mated to C57BL/6 males. Fertilized one-cell-stage embryos were collected from the oviducts, and the Cas9-encoding mRNA (100 ng/µl) and gRNA (50 ng/µl) were injected into the cytoplasm as described previously (Ma et al., 2014). The injected live embryos were transferred into the oviducts of pseudopregnant Institute of Cancer Research females 0.5 d after coitus, as described previously (Bando et al., 2019). The expression of Gnaq in the resulting tail fibroblast was analyzed by Western blot using protein-specific antibodies. Male pups harboring the desired mutation were mated to C57BL/6 female mice and tested for germline transmission of the mutated gene. Heterozygotic mice were intercrossed to generate homozygotic GNAQ-deficient mice to be used for in vitro and in vivo assays.
Mammalian expression plasmids
To construct the mammalian expression vectors, cDNA fragments were amplified using the following primer sets: mPLCβ4_F and mPLCβ4_R for PLCβ4(WT); mGNAQ_F and mGNAQ_R for GNAQ(WT); mGNA11_F and mGNA11_R for GNA11(WT); mCD3ε_F and mCD3ε_R for CD3ε; mCD4_F and mCD4_R for CD4; mCD8α_F and mCD8α_R for CD8α; FlpO_F and FlpO_R for FlpO; and Lck_F and Lck_R for Lck (Table S1). The series of PLCβ4 deletion mutants were generated using the following primers: mPLCβ4(880)_F and mPLCβ4_R for PLCβ4(880–1,175); mPLCβ4(900)_F and mPLCβ4_R for PLCβ4(900–1,175); mPLCβ4(920)_F and mPLCβ4_R for PLCβ4(920–1,175); mPLCβ4(940)_F and mPLCβ4_R for PLCβ4(940–1,175); mPLCβ4(960)_F and mPLCβ4_R for PLCβ4(960–1,175); mPLCβ4(T886A)_F and mPLCβ4(T886A)_R for PLCβ4(T886A); mPLCβ4(T886A/S889A)_F and mPLCβ4(T886A/S889A)_R for PLCβ4(T886A/S889A); mPLCβ4(T886A/S890A)_F and mPLCβ4(T886A/S890A)_R for PLCβ4(T886A/S890A); and mPLCβ4(T886A/S891A)_F and mPLCβ4(T886A/S891A)_R for PLCβ4(T886A/S891A) (Table S1). The series of CD8α deletion mutants were generated using the following primers: mCD8α_F and mCD8α (Δ5)_R for CD8α(1–242); mCD8α_F and mCD8α (Δ10)_R for CD8α(1–237); mCD8α_F and mCD8α (Δ15)_R for CD8α(1–232); mCD8α_F and mCD8α (Δ20)_R for CD8α(1–227); and mCD8α_F and mCD8α (Δ25)_R for CD8α(1–222) (Table S1). cDNA fragments encoding each protein were ligated into pcDNA vector for the C-terminal Flag-tagged, HA-tagged, or V5-tagged proteins. The sequences of all of the constructs were confirmed by sequencing analyses with an ABI PRISM Genetic Analyzer 3130xl (PE Applied Biosystems).
T cell proliferation assays
Purified CD8+ T cells and CD4+ T cells were labeled with 5 mM CFSE (Invitrogen). 1 × 105 T cells were stimulated with coated 5 µg/ml anti-CD3ε and soluble 2 µg/ml anti-CD28 antibodies for 72 h. The intensity of CFSE was analyzed using a FACSVerse (BD Biosciences).
Coculture experiments
WT or PLCβ4-deficient BMDCs were treated with 0.1 nM OVA257-264 peptide (TS5001-P; MBL) for 3 h. The cells were washed three times with PBS, fixed with 0.01% glutaraldehyde/PBS for 30 s, washed with PBS and RPMI 1640 supplemented with 10% FBS, and then cocultured in 96-well round-bottom plates with sorted WT or PLCβ4 OT-I T cells at various ratios for 72 h.
In vivo detection of Tetramer+ CD8+ or CD4+ T cells
WT and PLCβ4-deficient mice were intraperitoneally infected with γ-irradiated Pru T. gondii expressing p30-OVA or parental strain (1 × 106 per mouse) for 14 d. Mice were initially immunized with OVA (50 µg) and cyclic guanosine monophosphate–adenosine monophosphate (cGAMP; InvivoGen; 10 µg), followed by the second immunization 10 d after the first immunization. 7 d after the second immunization, spleens were taken and analyzed for presence of OVA-specific CD8+ T cells. Single-cell suspensions were prepared from the spleens of OVA-stimulated or infected mice; counted; stained with antibodies against CD3ε, CD8α, and OVA Tetramer-SIINFEKL (MBL; TS-5001-1C) or CD3ε, CD4, and OVA Tetramer-ISQAVHAAHAEINEAGR (MBL; TS-M710-1); and then acquired with a FACSVerse (BD Biosciences).
Measurement of T. gondii numbers by luciferase assay
To measure T. gondii survival rate, the cells were untreated or treated with IFN-γ (10 ng/ml) for 18 h and followed by the infection of Pru T. gondii expressing luciferase and OVA proteins (multiplicity of infection of 1) for 24 h. The infected cells were harvested and lysed with 100 µl of 1× passive lysis buffer (Promega) with sonication for 30 s. After centrifugation at 12,000 rpm for 10 min at 4°C, the luciferase activity from 5 µl of the supernatants was measured using the Dual-Luciferase Reporter Assay System (Promega) by GLOMAX20/20 luminometer (Promega). The data are presented as the percentage in T. gondii survival rate and relative to unstimulated cells.
Retroviral transduction of primary T cells
Retroviral supernatants were produced by transfecting Plate-E cells with pMRX-IRES-eGFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. PLCβ4, GNAQ, and GNA11 were cloned into pMRX-IRES-eGFP. Primary T cells were isolated from spleens of WT and PLCβ4-deficient mice. Purified T cells were activated with coated anti-CD3ε and soluble anti-CD28 antibodies for 48 h. Activated T cell pellet was resuspended in the retroviral supernatant at 1.5 × 106 cells/ml with 20 ng/ml recombinant murine IL-2. 1 ml of cell suspension was added to each well of a 24-well plate. The cell suspension was centrifuged at 2,000 g for 90 min at 32°C with no brake. After centrifugation, 1 ml of fresh RPMI with IL-2 (20 ng/ml) was added. After 12 h, medium was changed with RPMI containing IL-2 (20 ng/ml).
Intracellular cytokine staining
The intracellular expression of IFN-γ and TNF-α in CD8+ T cells was analyzed using the Cytofix/Cytoperm Kit Plus (with Golgistop; BD Biosciences) according to the manufacturer’s instructions. In brief, T cells obtained from the spleen were stimulated by incubation with coated anti-CD3ε and soluble anti-CD28 antibodies for 72 h and Golgistop in complete RPMI 1640 at 37°C for 4 h. Surface staining was performed with PeCy7-conjugated anti-CD8 for 20 min at 4°C. After Fix/Perm treatment for 20 min, intracellular cytokine staining was performed with Alexa Fluor 647–conjugated anti–IFN-γ and FITC–TNF-α for 20 min. Data were acquired using a FACS Verse and analyzed using FlowJo software.
Western blot analysis and immunoprecipitation
293T cells and primary CD8+ T cells were lysed in a lysis buffer (1% Nonidet P-40, 150 mM NaCl, and 20 mM Tris-HCl, pH 7.5) containing a protease inhibitor cocktail and phosphatase inhibitor (Nacalai Tesque). The cell lysates were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. For immunoprecipitation, cell lysates were precleared with protein G–Sepharose (GE Healthcare) for 2 h and then incubated with protein G–Sepharose containing 1 µg of indicated antibodies for 12 h with rotation at 4°C. The immunoprecipitants were washed four times with lysis buffer and eluted by boiling with Laemmli sample buffer. The eluates were separated by SDS-PAGE, transferred to polyvinylidene fluoride membranes, and subjected to Western blotting using the indicated antibodies, as described above. To isolate the samples from mesenteric lymph node, spleens, and brains, the tissues and organs were mechanistically triturated followed by single separation. Cell numbers were counted and adjusted to 2 × 106 cells/lane (Fig. S1 D).
Generation of recombinant PLCβ4 and CD8α using cell-free system
His-tagged recombinant PLCβ4-HA and CD8α-Flag were expressed with wheat germ cell-free system (CellFree Sciences) and purified with a Nickel-Sepharose 6 fast flow (GE Healthcare), as previously reported (Tsuboi et al., 2010). Protein purity was evaluated by SDS-PAGE and Coomassie Brilliant Blue staining. The purified PLCβ4-HA and/or CD8α_Flag proteins (100 ng/each) were resuspended in the same lysis buffer used in the immunoprecipitation, which was performed as described above.
MS
293T cells transiently cotransfected with HA-tagged PLCβ4 WT or various PLCβ4 mutants and Flag-tagged GNAQ (Q209L) were lysed in RIPA buffer (20 mM Hepes-NaOH, pH 7.5, 1 mM EGTA, 1 mM MgCl2, 150 mM NaCl, 0.25% sodium deoxycholate, 0.15% SDS, and 1% NP-40) containing a protease inhibitor cocktail and phosphatase inhibitors (Nacalai Tesque). After centrifugation at 5,000 rpm for 15 min at 4°C, the supernatants were incubated with a 20-µl slurry of anti–HA-agarose (Sigma-Aldrich) for 3 h at 4°C. The beads were washed four times with RIPA buffer and then twice with 50 mM ammonium bicarbonate. Proteins on the beads were digested by adding 200 ng trypsin/Lys-C mix (Promega) at 37°C for 16 h. The digests were reduced, alkylated, acidified, and desalted using GL-Tip SDB (GL Sciences). The eluates were evaporated in a SpeedVac concentrator and dissolved in 0.1% trifluoroacetic acid and 3% acetonitrile (ACN). Liquid chromatography-MS/MS analysis of the resultant peptides was performed on an EASY-nLC 1200 UHPLC connected to an Orbitrap Fusion mass spectrometer through a nanoelectrospray ion source (Thermo Fisher Scientific). The peptides were separated on a 75-µm inner diameter × 150-mm C18 reversed-phase column (Nikkyo Technos) with a linear 4–32% ACN gradient for 0–100 min, followed by an increase to 80% ACN for 10 min. The mass spectrometer was operated in data-dependent acquisition mode with a maximum duty cycle of 3 s. MS1 spectra were measured with a resolution of 60,000, an automatic gain control (AGC) target of 4 × 105, and a mass range from 350 to 1,500 m/z. High-energy collisional dissociation MS/MS spectra were acquired in the Orbitrap with a resolution of 30,000, an AGC target of 5 × 104, an isolation window of 1.6 m/z, a maximum injection time of 54 ms, and a normalized collision energy of 30. Then collision-induced dissociation MS/MS spectra were acquired in the Orbitrap with a resolution of 30,000, an AGC target of 5 × 104, an isolation window of 1.6 m/z, a maximum injection time of 54 ms, and a normalized collision energy of 35. Dynamic exclusion was set to 15 s. Raw data were directly analyzed against the SwissProt database restricted to Homo sapiens supplemented with mouse PLCβ4 and GNAQ protein sequences using Proteome Discoverer version 2.4 (Thermo Fisher Scientific) with Sequest HT search engine. The search parameters were as follows: (a) trypsin as an enzyme with up to two missed cleavages; (b) precursor mass tolerance of 10 ppm; (c) fragment mass tolerance of 0.02 D; (d) carbamidomethylation of cysteine as a fixed modification; and (e) acetylation of protein N-terminus, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine as variable modifications. Peptides were filtered at a false discovery rate of 1% using the percolator node.
Several selected peptides of mouse PLCβ4 (WT and various mutants) were measured by parallel reaction monitoring, an MS/MS-based targeted quantification method using high-resolution MS. Targeted high-energy collision dissociation and collision-induced dissociation MS/MS scans were acquired by a time-scheduled inclusion list at a resolution of 30,000, an AGC target of 5 × 104, an isolation window of 1.6 m/z, a maximum injection time of 300 ms, and a normalized collision energy of 30. Time alignment and relative quantification of the transitions were performed using Skyline software.
Measurement of cytokines by ELISA
The concentration of IFN-γ was measured by ELISA according to the manufacturer’s protocol (eBioscience). The PIP2 levels in flow cytometry–sorted WT and PLCβ4-deficient CD8+ T cells (105 each/50 µl PBS) was measured by mouse PIP2 ELISA Kit (MyBiosource).
Measurement of IP1 as indicator of IP3 production
Purified CD8+ T cells from WT and PLCβ4-deficient mice were stimulated with indicated doses of anti-CD3 for 1 h in the presence of 50 mM LiCl. Following lysis, the supernatant was transferred into an ELISA plate, and the IP1 levels were determined by addition of ELISA reagents according to the manufacturer’s protocol (Cisbio-US).
Quantitative RT-PCR
Total RNA was extracted and cDNA was synthesized using Verso Reverse transcription (Thermo Fisher Scientific). Real-time PCR was performed with a CFX connect real-time PCR system (Bio-Rad) using the Go-Taq real-time PCR system (Promega). The values were normalized to the amount of GAPDH in each sample. The following primer sets were used: IL-2_qpF and IL-2_qpR for IL-2; IFN-γ_qpF and IFN-γ_qpR for Ifng; GAPDH_qpF and GAPDH_qpR for Gapdh; Gzmb_qpF and Gzmb_qpR for Gzmb; PLCβ4_qpF and PLCβ4_qpR for Plcb4; GNAQ_qpF and GNAQ_qpR for Gnaq; GNA11_qpF and GNA11_qpR for Gna11; GNA14_qpF and GNA14_qpR for Gna14; and GNA15_qpF and GNA15_qpR for Gna15 (Table S1).
In vivo imaging analysis
Mice were intraperitoneally infected with 105 freshly egressed tachyzoites expressing luciferase resuspended in 100 µl PBS. Bioluminescence was assessed on the indicated days after infection. For the detection of bioluminescence emission, mice were intraperitoneally injected with 3 mg D-luciferin (Promega) in 200 µl PBS, maintained for 5 min to allow for adequate dissemination of luciferin, and subsequently anaesthetized with isoflurane (Dainippon Sumitomo Pharma). 10 min after injection of D-luciferin, photonic emissions were detected using an in vivo imaging system (IVIS-Spectrum; Xenogen) and Living Image software (Xenogen).
Calcium flux analysis
Purified CD8+ T cells from the spleens of WT and PLCβ4 KO mice were incubated with 4 µM Fluo-8 (Abcam) in RPMI 1640 at 37°C, 5% incubator for 1 h. The cells were stained with PE-cy7 anti-CD8 and anti-CD3e (14C11) in 100 µl PBS (2% FBS) for 15 min. After washing, the cells were resuspended in 500 µl FACS buffer (PBS, 1% BSA, and 500 µM CaCl2) and run on an Aria III (BD). Baseline data were collected for 1 min, anti-hamster IgG was added to a final concentration of 10 µg/ml to cross-link to TCR, and data were then collected by the Aria III for another 4 min.
Isolation of splenic naive CD4+ cells and in vitro Th1 differentiation
For FACS sorting, splenocytes from WT or PLCβ4-deficient mice were stained with PE-conjugated anti-CD4, PerCP/Cy5.5-conjugated anti-CD62L, Pacific Blue–conjugated anti-CD25, and PE-Cy7–conjugated anti-CD44. Naive CD4+ T cells were sorted using a FACSAria III for CD4+CD62LhighCD25−CD44low. The purity of the sorted cells was routinely >98%. For Th1 differentiation, naive CD4+ T cells from WT or PLCβ4-deficient mice were cultured with plate-bound anti-CD3ε and anti-CD28 Abs in the presence of IL-12 (10 ng/ml) and anti-IL4 (1 µg/ml) at 2.5 × 105 cells/ml for 3 d.
Adoptive transfer of T cells
Polyclonal CD8+ T cells were isolated from the spleens of WT and PLCβ4 KO mice by Aria III (BD Biosciences). Isolated CD8+ T cells (1 × 106) were injected intravenously into PLCβ4 KO mice 18 h before infection with T. gondii.
Melanoma mouse models
B16F10 cells were washed with PBS three times. B16F10 cells (2 × 105 cells in 200 µl) were intravenously injected in the tail veins of mice. The mice survival rate was monitored every day. To study tumor growth and metastasis, lungs were harvested on the indicated following days, and tumor numbers on lungs were counted.
Statistical analysis
Three points in all graphs represent three means derived from three independent experiments (three biological replicates). All statistical analyses were performed using Prism 7 (GraphPad). The statistical significance of differences in mean values was analyzed by using an unpaired two-tailed Student’s t test. P values <0.05 were considered to be statistically significant. The statistical significance of differences in survival times of mice between two groups was analyzed by Kaplan–Meier survival analysis log-rank test.
Online supplemental material
Fig. S1 shows the strategy of PLCβ4 gene-trapped mice and floxed mice. Fig. S2 shows comparable CD4+ and CD8+ T cell cellularity in thymus and spleens in WT and PLCβ4-deficient mice. Fig. S3 shows comparable composition of CD44 and CD62L of CD4+ or CD8+ T cells in WT and PLCβ4-deficient mice. Fig. S4 shows competitive binding of Lck and PLCβ4 with CD8α in vitro. Fig. S5 shows normal CD4+ activation in T. gondii–infected PLCβ4-deficient mice. Table S1 lists primers used in this study.
Supplementary Material
lists primers used in this study.
Acknowledgments
We thank Drs. A. Iwasaki (Yale University) and S. Yamamoto (Medical College of Oita) for helpful discussion. We thank M. Enomoto (Osaka University) for secretarial assistance.
This study was supported by the Japan Agency for Medical Research and Development: Research Program on Emerging and Re-emerging Infectious Diseases (JP20fk0108137 for M. Yamamoto), Japanese Initiative for Progress of Research on Infectious Diseases for Global Epidemic (JP20wm0325010 for M. Yamamoto), and Strategic International Collaborative Research Program (JP19jm0210067 for M. Yamamoto); the Ministry of Education, Culture, Sports, Science and Technology: Grant-in-Aid for Transformative Research Area (B) (20H05771 for M. Yamamoto), for Scientific Research on Innovative Areas (19H04809 for M. Yamamoto), for Scientific Research (B) (18KK0226 for M. Yamamoto and 18H02642 for M. Sasai), and for Scientific Research (A) (19H00970 for M. Yamamoto); Joint Usage and Joint Research Programs of the Institute of Advanced Medical Sciences, Tokushima University for M. Yamamoto; Takeda Science Foundation for M. Yamamoto and M. Sasai; Mochida Memorial Foundation for Medical and Pharmaceutical Research for M. Yamamoto and M. Sasai; Uehara Memorial Foundation for M. Yamamoto and M. Sasai; Naito Foundation for M. Yamamoto and M. Sasai; Astellas Foundation for Research on Metabolic Disorders for M. Sasai; and Research Foundation for Microbial Diseases of Osaka University for M. Yamamoto.
Author contributions: M. Sasai, J.S. Ma, M. Okamoto, K. Nishino, H. Nagaoka, E. Takashima, H. Kosako, and M. Yamamoto designed and performed experiments, analyzed the data, and prepared the figures; A. Pradipta, Y. Lee, and P.-G. Suh prepared research reagents and mice; M. Yamamoto conceived the project and supervised this research.
References
- Anderson, D.L., and Tsoukas C.D.. 1989. Cholera toxin inhibits resting human T cell activation via a cAMP-independent pathway. J. Immunol. 143:3647–3652. [PubMed] [Google Scholar]
- Artyomov, M.N., Lis M., Devadas S., Davis M.M., and Chakraborty A.K.. 2010. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl. Acad. Sci. USA. 107:16916–16921. 10.1073/pnas.1010568107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando, H., Pradipta A., Iwanaga S., Okamoto T., Okuzaki D., Tanaka S., Vega-Rodríguez J., Lee Y., Ma J.S., Sakaguchi N., et al. 2019. CXCR4 regulates Plasmodium development in mouse and human hepatocytes. J. Exp. Med. 216:1733–1748. 10.1084/jem.20182227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, T.A., and Amir E.. 2017. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br. J. Cancer. 117:451–460. 10.1038/bjc.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd, J.C. 2009. Toxoplasma gondii: 25 years and 25 major advances for the field. Int. J. Parasitol. 39:935–946. 10.1016/j.ijpara.2009.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno, C., Lemke C.D., Criado G., Baroja M.L., Ferguson S.S., Rahman A.K., Tsoukas C.D., McCormick J.K., and Madrenas J.. 2006. Bacterial superantigens bypass Lck-dependent T cell receptor signaling by activating a Galpha11-dependent, PLC-beta-mediated pathway. Immunity. 25:67–78. 10.1016/j.immuni.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Casciotti, L., Ely K.H., Williams M.E., and Khan I.A.. 2002. CD8(+)-T-cell immunity against Toxoplasma gondii can be induced but not maintained in mice lacking conventional CD4(+) T cells. Infect. Immun. 70:434–443. 10.1128/IAI.70.2.434-443.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciarelli, C., Hohman R.J., Atkinson T.P., Gusovsky F., and Weissman A.M.. 1992. Evidence for GTP-binding protein involvement in the tyrosine phosphorylation of the T cell receptor zeta chain. J. Biol. Chem. 267:14527–14530. 10.1016/S0021-9258(18)42068-6 [DOI] [PubMed] [Google Scholar]
- Chan, I.T., Limmer A., Louie M.C., Bullock E.D., Fung-Leung W.P., Mak T.W., and Loh D.Y.. 1993. Thymic selection of cytotoxic T cells independent of CD8 alpha-Lck association. Science. 261:1581–1584. 10.1126/science.8372352 [DOI] [PubMed] [Google Scholar]
- Clements, J.L., Yang B., Ross-Barta S.E., Eliason S.L., Hrstka R.F., Williamson R.A., and Koretzky G.A.. 1998. Requirement for the leukocyte-specific adapter protein SLP-76 for normal T cell development. Science. 281:416–419. 10.1126/science.281.5375.416 [DOI] [PubMed] [Google Scholar]
- Combe, C.L., Curiel T.J., Moretto M.M., and Khan I.A.. 2005. NK cells help to induce CD8(+)-T-cell immunity against Toxoplasma gondii in the absence of CD4(+) T cells. Infect. Immun. 73:4913–4921. 10.1128/IAI.73.8.4913-4921.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, A.H., Lo W.L., and Weiss A.. 2018. TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem. Sci. 43:108–123. 10.1016/j.tibs.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin, M.L., and Choudhuri K.. 2016. Signaling and Polarized Communication Across the T Cell Immunological Synapse. Annu. Rev. Cell Dev. Biol. 32:303–325. 10.1146/annurev-cellbio-100814-125330 [DOI] [PubMed] [Google Scholar]
- Eichmann, K., Boyce N.W., Schmidt-Ullrich R., and Jönsson J.I.. 1989. Distinct functions of CD8(CD4) are utilized at different stages of T-lymphocyte differentiation. Immunol. Rev. 109:39–75. 10.1111/j.1600-065X.1989.tb00019.x [DOI] [PubMed] [Google Scholar]
- Evanko, D.S., Thiyagarajan M.M., and Wedegaertner P.B.. 2000. Interaction with Gbetagamma is required for membrane targeting and palmitoylation of Galpha(s) and Galpha(q). J. Biol. Chem. 275:1327–1336. 10.1074/jbc.275.2.1327 [DOI] [PubMed] [Google Scholar]
- Fridman, W.H., Pagès F., Sautès-Fridman C., and Galon J.. 2012. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- Fu, G., Chen Y., Yu M., Podd A., Schuman J., He Y., Di L., Yassai M., Haribhai D., North P.E., et al. 2010. Phospholipase Cgamma1 is essential for T cell development, activation, and tolerance. J. Exp. Med. 207:309–318. 10.1084/jem.20090880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G., Rybakin V., Brzostek J., Paster W., Acuto O., and Gascoigne N.R.. 2014. Fine-tuning T cell receptor signaling to control T cell development. Trends Immunol. 35:311–318. 10.1016/j.it.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung-Leung, W.P., Louie M.C., Limmer A., Ohashi P.S., Ngo K., Chen L., Kawai K., Lacy E., Loh D.Y., and Mak T.W.. 1993. The lack of CD8 alpha cytoplasmic domain resulted in a dramatic decrease in efficiency in thymic maturation but only a moderate reduction in cytotoxic function of CD8+ T lymphocytes. Eur. J. Immunol. 23:2834–2840. 10.1002/eji.1830231117 [DOI] [PubMed] [Google Scholar]
- Gaud, G., Lesourne R., and Love P.E.. 2018. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18:485–497. 10.1038/s41577-018-0020-8 [DOI] [PubMed] [Google Scholar]
- Gilman, A.G. 1984. G proteins and dual control of adenylate cyclase. Cell. 36:577–579. 10.1016/0092-8674(84)90336-2 [DOI] [PubMed] [Google Scholar]
- Golstein, P., and Griffiths G.M.. 2018. An early history of T cell-mediated cytotoxicity. Nat. Rev. Immunol. 18:527–535. 10.1038/s41577-018-0009-3 [DOI] [PubMed] [Google Scholar]
- Grabocka, E., and Wedegaertner P.B.. 2005. Functional consequences of G alpha 13 mutations that disrupt interaction with p115RhoGEF. Oncogene. 24:2155–2165. 10.1038/sj.onc.1208414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, J.D., and Cantrell D.A.. 1991. An analysis of the role of guanine nucleotide binding proteins in antigen receptor/CD3 antigen coupling to phospholipase C. J. Immunol. 146:2102–2107. [PubMed] [Google Scholar]
- Horejsí, V., Zhang W., and Schraven B.. 2004. Transmembrane adaptor proteins: organizers of immunoreceptor signalling. Nat. Rev. Immunol. 4:603–616. 10.1038/nri1414 [DOI] [PubMed] [Google Scholar]
- Howden, A.J.M., Hukelmann J.L., Brenes A., Spinelli L., Sinclair L.V., Lamond A.I., and Cantrell D.A.. 2019. Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 20:1542–1554. 10.1038/s41590-019-0495-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden, J.B., Shoback D.M., Pattison G., and Stobo J.D.. 1986. Cholera toxin inhibits the T-cell antigen receptor-mediated increases in inositol trisphosphate and cytoplasmic free calcium. Proc. Natl. Acad. Sci. USA. 83:5673–5677. 10.1073/pnas.83.15.5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway, C.A. Jr. 1992. The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu. Rev. Immunol. 10:645–674. 10.1146/annurev.iy.10.040192.003241 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Lyubarsky A., Dodd R., Vardi N., Pugh E., Baylor D., Simon M.I., and Wu D.. 1996. Phospholipase C beta 4 is involved in modulating the visual response in mice. Proc. Natl. Acad. Sci. USA. 93:14598–14601. 10.1073/pnas.93.25.14598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius, M., Maroun C.R., and Haughn L.. 1993. Distinct roles for CD4 and CD8 as co-receptors in antigen receptor signalling. Immunol. Today. 14:177–183. 10.1016/0167-5699(93)90282-P [DOI] [PubMed] [Google Scholar]
- Kadamur, G., and Ross E.M.. 2013. Mammalian phospholipase C. Annu. Rev. Physiol. 75:127–154. 10.1146/annurev-physiol-030212-183750 [DOI] [PubMed] [Google Scholar]
- Kaech, S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano, M., Hashimoto K., Watanabe M., Kurihara H., Offermanns S., Jiang H., Wu Y., Jun K., Shin H.S., Inoue Y., et al. 1998. Phospholipase cbeta4 is specifically involved in climbing fiber synapse elimination in the developing cerebellum. Proc. Natl. Acad. Sci. USA. 95:15724–15729. 10.1073/pnas.95.26.15724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, T., and Xiao W.. 2013. Phospholipase C-β in immune cells. Adv. Biol. Regul. 53:249–257. 10.1016/j.jbior.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Jun K.S., Lee S.B., Kang N.G., Min D.S., Kim Y.H., Ryu S.H., Suh P.G., and Shin H.S.. 1997. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature. 389:290–293. 10.1038/38508 [DOI] [PubMed] [Google Scholar]
- Kim, P.W., Sun Z.Y., Blacklow S.C., Wagner G., and Eck M.J.. 2003. A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8. Science. 301:1725–1728. 10.1126/science.1085643 [DOI] [PubMed] [Google Scholar]
- Lee, C.W., Lee K.H., Lee S.B., Park D., and Rhee S.G.. 1994. Regulation of phospholipase C-beta 4 by ribonucleotides and the alpha subunit of Gq. J. Biol. Chem. 269:25335–25338. 10.1016/S0021-9258(18)47252-3 [DOI] [PubMed] [Google Scholar]
- Lengagne, R., Graff-Dubois S., Garcette M., Renia L., Kato M., Guillet J.G., Engelhard V.H., Avril M.F., Abastado J.P., and Prévost-Blondel A.. 2008. Distinct role for CD8 T cells toward cutaneous tumors and visceral metastases. J. Immunol. 180:130–137. 10.4049/jimmunol.180.1.130 [DOI] [PubMed] [Google Scholar]
- Lu, F., Huang S., and Kasper L.H.. 2009. The temperature-sensitive mutants of Toxoplasma gondii and ocular toxoplasmosis. Vaccine. 27:573–580. 10.1016/j.vaccine.2008.10.090 [DOI] [PubMed] [Google Scholar]
- Lyon, A.M., and Tesmer J.J.. 2013. Structural insights into phospholipase C-β function. Mol. Pharmacol. 84:488–500. 10.1124/mol.113.087403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.S., Sasai M., Ohshima J., Lee Y., Bando H., Takeda K., and Yamamoto M.. 2014. Selective and strain-specific NFAT4 activation by the Toxoplasma gondii polymorphic dense granule protein GRA6. J. Exp. Med. 211:2013–2032. 10.1084/jem.20131272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, T.J.K., Kishihara K., Siderovski D.P., van Ewijk W., Narendran A., Timms E., Wakeham A., Paige C.J., Hartmann K.U., Veillette A., et al. 1992. Profound block in thymocyte development in mice lacking p56lck. Nature. 357:161–164. 10.1038/357161a0 [DOI] [PubMed] [Google Scholar]
- Negishi, I., Motoyama N., Nakayama K., Nakayama K., Senju S., Hatakeyama S., Zhang Q., Chan A.C., and Loh D.Y.. 1995. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature. 376:435–438. 10.1038/376435a0 [DOI] [PubMed] [Google Scholar]
- Ngai, J., Methi T., Andressen K.W., Levy F.O., Torgersen K.M., Vang T., Wettschureck N., and Taskén K.. 2008. The heterotrimeric G-protein alpha-subunit Galphaq regulates TCR-mediated immune responses through an Lck-dependent pathway. Eur. J. Immunol. 38:3208–3218. 10.1002/eji.200838195 [DOI] [PubMed] [Google Scholar]
- O’Shea, J.J., Urdahl K.B., Luong H.T., Chused T.M., Samelson L.E., and Klausner R.D.. 1987. Aluminum fluoride induces phosphatidylinositol turnover, elevation of cytoplasmic free calcium, and phosphorylation of the T cell antigen receptor in murine T cells. J. Immunol. 139:3463–3469. [PubMed] [Google Scholar]
- Parnes, J.R. 1989. Molecular biology and function of CD4 and CD8. Adv. Immunol. 44:265–311. 10.1016/S0065-2776(08)60644-6 [DOI] [PubMed] [Google Scholar]
- Peng, Y.W., Rhee S.G., Yu W.P., Ho Y.K., Schoen T., Chader G.J., and Yau K.W.. 1997. Identification of components of a phosphoinositide signaling pathway in retinal rod outer segments. Proc. Natl. Acad. Sci. USA. 94:1995–2000. 10.1073/pnas.94.5.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazzotti, G., Faenza I., Gaboardi G.C., Piazzi M., Bavelloni A., Fiume R., Manzoli L., Martelli A.M., and Cocco L.. 2008. Catalytic activity of nuclear PLC-beta(1) is required for its signalling function during C2C12 differentiation. Cell. Signal. 20:2013–2021. 10.1016/j.cellsig.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Rao, A., and Hogan P.G.. 2009. Calcium signaling in cells of the immune and hematopoietic systems. Immunol. Rev. 231:5–9. 10.1111/j.1600-065X.2009.00823.x [DOI] [PubMed] [Google Scholar]
- Rhee, S.G. 2001. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70:281–312. 10.1146/annurev.biochem.70.1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann, J.C., Geiger R., Hornburg D., Wolf T., Kveler K., Jarrossay D., Sallusto F., Shen-Orr S.S., Lanzavecchia A., Mann M., and Meissner F.. 2017. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 18:583–593. 10.1038/ni.3693 [DOI] [PubMed] [Google Scholar]
- Rudd, C.E., Trevillyan J.M., Dasgupta J.D., Wong L.L., and Schlossman S.F.. 1988. The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. Proc. Natl. Acad. Sci. USA. 85:5190–5194. 10.1073/pnas.85.14.5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson, L.E. 2002. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 20:371–394. 10.1146/annurev.immunol.20.092601.111357 [DOI] [PubMed] [Google Scholar]
- Samelson, L.E. 2011. Immunoreceptor signaling. Cold Spring Harb. Perspect. Biol. 3:a011510. 10.1101/cshperspect.a011510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson, L.E., and Klausner R.D.. 1988. The T-cell antigen receptor. Structure and mechanism of activation. Ann. N. Y. Acad. Sci. 540(1 Advances in N):1–3. 10.1111/j.1749-6632.1988.tb27045.x [DOI] [PubMed] [Google Scholar]
- Sasai, M., Pradipta A., and Yamamoto M.. 2018. Host immune responses to Toxoplasma gondii. Int. Immunol. 30:113–119. 10.1093/intimm/dxy004 [DOI] [PubMed] [Google Scholar]
- Shaw, A.S., Chalupny J., Whitney J.A., Hammond C., Amrein K.E., Kavathas P., Sefton B.M., and Rose J.K.. 1990. Short related sequences in the cytoplasmic domains of CD4 and CD8 mediate binding to the amino-terminal domain of the p56lck tyrosine protein kinase. Mol. Cell. Biol. 10:1853–1862. 10.1128/MCB.10.5.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrcka, A.V., and Sternweis P.C.. 1993. Regulation of purified subtypes of phosphatidylinositol-specific phospholipase C beta by G protein alpha and beta gamma subunits. J. Biol. Chem. 268:9667–9674. 10.1016/S0021-9258(18)98401-2 [DOI] [PubMed] [Google Scholar]
- Stanners, J., Kabouridis P.S., McGuire K.L., and Tsoukas C.D.. 1995. Interaction between G proteins and tyrosine kinases upon T cell receptor.CD3-mediated signaling. J. Biol. Chem. 270:30635–30642. 10.1074/jbc.270.51.30635 [DOI] [PubMed] [Google Scholar]
- Suh, P.G., Park J.I., Manzoli L., Cocco L., Peak J.C., Katan M., Fukami K., Kataoka T., Yun S., and Ryu S.H.. 2008. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41:415–434. 10.5483/BMBRep.2008.41.6.415 [DOI] [PubMed] [Google Scholar]
- Telfer, J.C., and Rudd C.E.. 1991. A 32-kD GTP-binding protein associated with the CD4-p56lck and CD8-p56lck T cell receptor complexes. Science. 254:439–441. 10.1126/science.1925604 [DOI] [PubMed] [Google Scholar]
- Thomsen, W., Frazer J., and Unett D.. 2005. Functional assays for screening GPCR targets. Curr. Opin. Biotechnol. 16:655–665. [DOI] [PubMed] [Google Scholar]
- Tsuboi, T., Takeo S., Sawasaki T., Torii M., and Endo Y.. 2010. An efficient approach to the production of vaccines against the malaria parasite. Methods Mol. Biol. 607:73–83. 10.1007/978-1-60327-331-2_8 [DOI] [PubMed] [Google Scholar]
- Turner, J.M., Brodsky M.H., Irving B.A., Levin S.D., Perlmutter R.M., and Littman D.R.. 1990. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 60:755–765. 10.1016/0092-8674(90)90090-2 [DOI] [PubMed] [Google Scholar]
- Veillette, A., Bookman M.A., Horak E.M., and Bolen J.B.. 1988. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 55:301–308. 10.1016/0092-8674(88)90053-0 [DOI] [PubMed] [Google Scholar]
- Veillette, A., Zúñiga-Pflücker J.C., Bolen J.B., and Kruisbeek A.M.. 1989. Engagement of CD4 and CD8 expressed on immature thymocytes induces activation of intracellular tyrosine phosphorylation pathways. J. Exp. Med. 170:1671–1680. 10.1084/jem.170.5.1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisinne, G., Kersse K., Chaoui K., Lu L., Chaix J., Zhang L., Goncalves Menoita M., Girard L., Ounoughene Y., Wang H., et al. 2019. Quantitative interactomics in primary T cells unveils TCR signal diversification extent and dynamics. Nat. Immunol. 20:1530–1541. 10.1038/s41590-019-0489-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M., Nakamura M., Sato K., Kano M., Simon M.I., and Inoue Y.. 1998. Patterns of expression for the mRNA corresponding to the four isoforms of phospholipase Cbeta in mouse brain. Eur. J. Neurosci. 10:2016–2025. 10.1046/j.1460-9568.1998.00213.x [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Ma J.S., Mueller C., Kamiyama N., Saiga H., Kubo E., Kimura T., Okamoto T., Okuyama M., Kayama H., et al. 2011. ATF6beta is a host cellular target of the Toxoplasma gondii virulence factor ROP18. J. Exp. Med. 208:1533–1546. 10.1084/jem.20101660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska, R. 1994. The CD8 coreceptor revisited: one chain good, two chains better. Immunity. 1:243–246. 10.1016/1074-7613(94)90075-2 [DOI] [PubMed] [Google Scholar]
- Zhang, W., Sommers C.L., Burshtyn D.N., Stebbins C.C., DeJarnette J.B., Trible R.P., Grinberg A., Tsay H.C., Jacobs H.M., Kessler C.M., et al. 1999. Essential role of LAT in T cell development. Immunity. 10:323–332. 10.1016/S1074-7613(00)80032-1 [DOI] [PubMed] [Google Scholar]
- Zhou, J., Stanners J., Kabouridis P., Han H., and Tsoukas C.D.. 1998. Inhibition of TCR/CD3-mediated signaling by a mutant of the hematopoietically expressed G16 GTP-binding protein. Eur. J. Immunol. 28:1645–1655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primers used in this study.