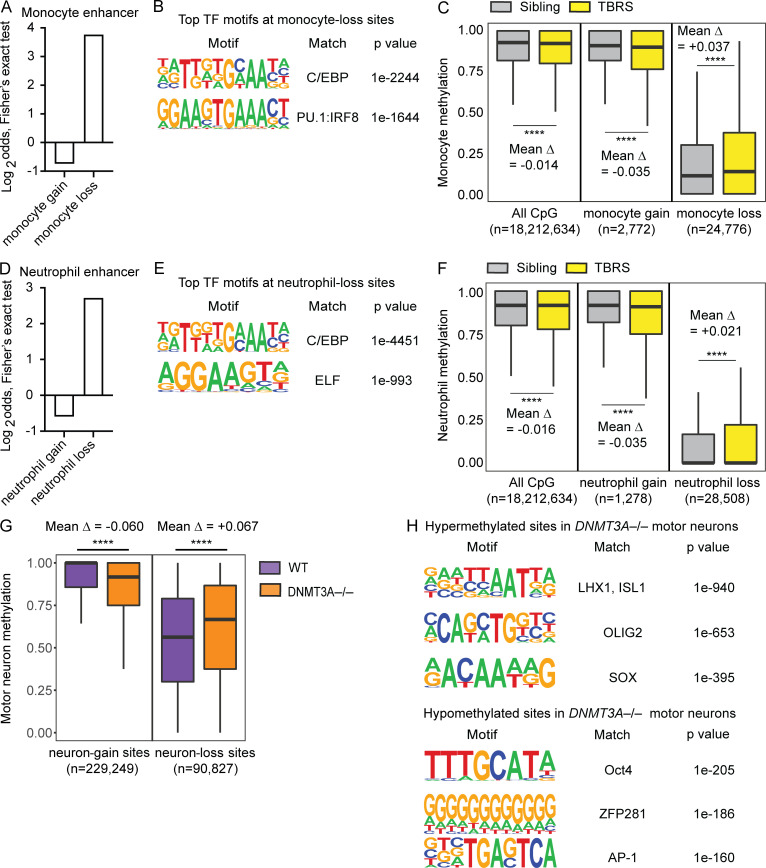
Figure 5.
DNMT3A defects cause DNA hypermethylation at cell-type–specific enhancers in primary myeloid cells and hESC-derived neurons. Results of analyzing publicly available datasets: A–F (Spencer et al., 2017) and G and H (Ziller et al., 2018). (A) Two-sided Fisher’s exact test results (Log2odds) for enrichment/depletion of the monocyte enhancer chromatin state in monocyte-gain and monocyte-loss sites (P < 0.05 for both tests). (B) Top two significant TF binding motifs enriched near (±100 bp) the monocyte-loss sites. (C) Methylation levels of monocytes from a TBRS patient and his sibling are compared at all examined CpG, monocyte-gain, and monocyte-loss sites. The number of CpG sites included in each analysis and mean differences in the methylation level (Δ) are shown. ****, P < 0.0001 by paired t test. (D) Same as A for neutrophils. (E) Top two significant TF binding motifs enriched near (±100 bp) the neutrophil-loss sites. (F) Same as C for neutrophils. (G) Methylation levels of motor neurons differentiated from WT or DNMT3A−/− HUES64-hESCs are compared at neuron-gain and neuron-loss sites. The number of CpG sites included in each analysis and mean differences in the methylation level (Δ) are shown. ****, P < 0.0001 by paired t test. (H) Top three significant TF binding motifs enriched near (±100 bp) the hyper- or hypomethylated sites in DNMT3A−/− motor neurons.