Figure 1.
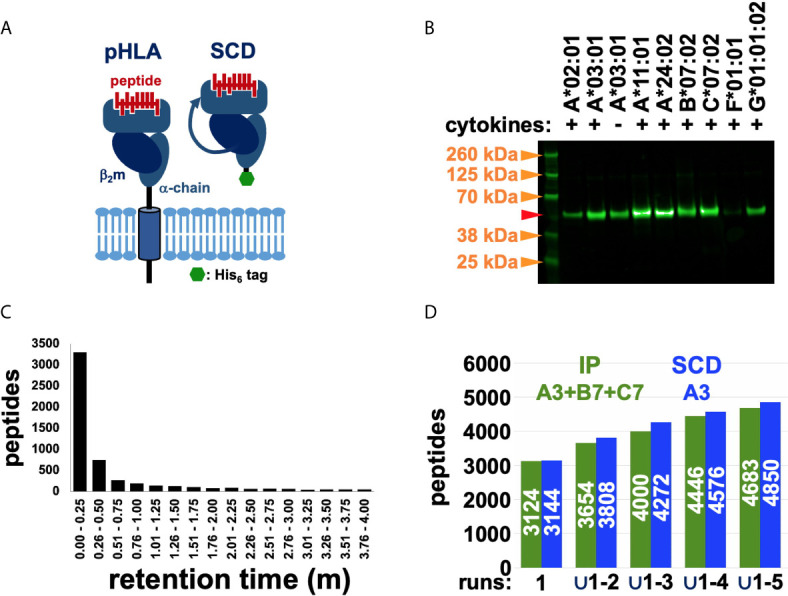
Using the SCD construct to recover and MS sequence HLA-restricted peptides. (A) Schematic representations highlight the differences between native, cell-surface pHLA and the engineered, secreted SCD construct. (B) Western blot analysis of SCD expression levels across alleles confirmed secretion of all tested SCDs, though at variable levels. Transduction, tissue culture, and Western conditions were closely matched across samples to allow meaningful comparisons of expression levels. The A3 SCD was tested with and without added cytokines (IFNγ, TNFα). Molecular weight markers (orange arrows) are indicated; the red arrow marks the expected PAGE mobility of an SCD with normal, expected N-glycosylation. (C) Chromatography retention times for nested peptides are graphed. The y-axis shows total number of unique peptide sequences in nested peptide sets in a co-elution time range; retention time intervals are indicated along the x-axis. (D) Accumulation rates of unique peptide sequences across five experimental replicates from the IP (green) and SCD (blue) datasets are compared (the pan-HLA IP dataset spans the three alleles in the haplotype of the cells analyzed, A3, B7, C7, but only A3 SCD results are shown). The y-axis shows total number of unique peptide sequences (also indicated in white in the bars) and progressively larger unions of five replicate datasets are indicated along the x-axis.
