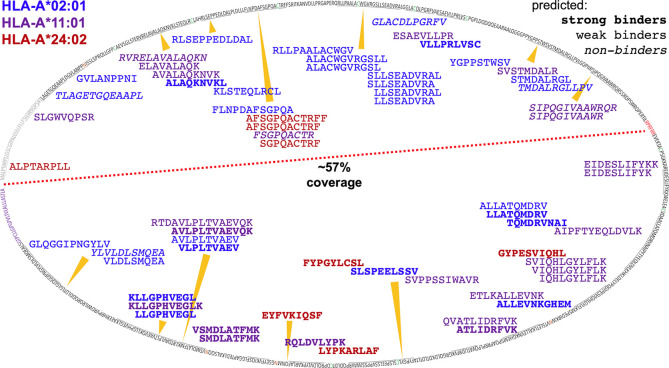
Figure 11.
Mapping ARTEMIS-identified peptides onto the MSLN sequence. The sequence of MSLN is shown mapped onto an oval. The leader peptide is colored gray, the furin cleavage site is colored red, the 4x potential N-glycan sites are colored orange, and the GPI signal sequence is colored purple. The red dashed line separates the MSLN proprotein sequence into MPF and MSLN moieties. ARTEMIS-identified peptides are shown inside the oval, approximately positioned by their location in the sequence, colored by cognate allele as indicated, with predicted binding/non-binding behavior by NetMHCpan shown in bold or italics as indicated. 57% of the MPF/MSLN sequence is presented as A2, A11, or A24 peptides identifiable via ARTEMIS. The MPF moiety of the MSLN fusion precursor proteins is highly expressed as a secreted protein via transduction in HEK293 cells, ten-fold or more higher in culture supernatants than SCDs, so may contaminate the isolated SCDs. However, observed coverage was comparable over the two moieties, including peptides from the leader sequence (which are not derived from a secreted contaminant but from endogenously expressed MPF/MSLN), suggesting that contamination was not a huge issue. 115 A2, A11, and A24 peptides were predicted to bind, using NetMHCpan, that were not observed by ARTEMIS. These peptides have not been shown to avoid excessive clutter.