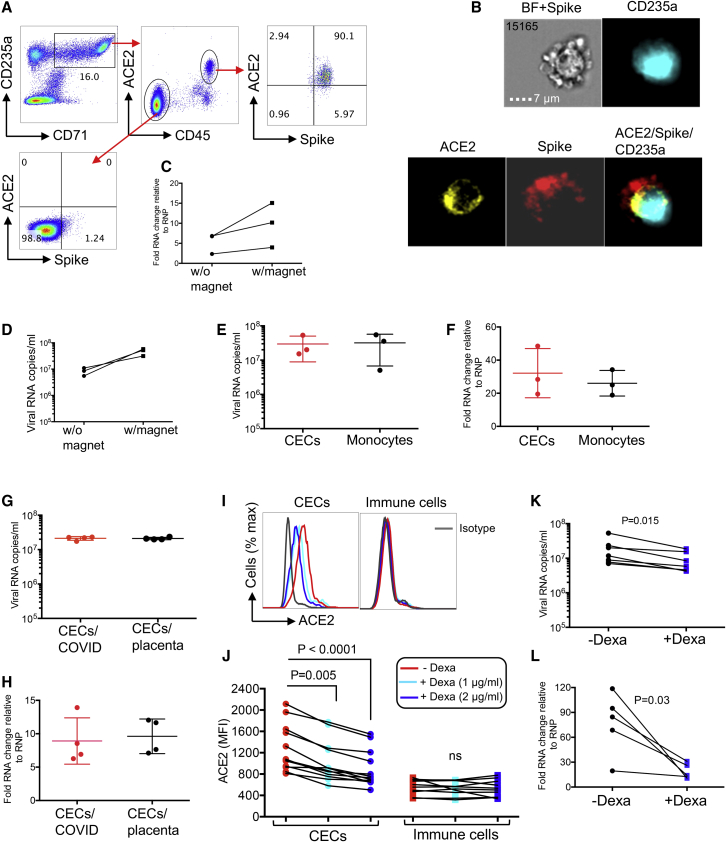
Figure 6.
CECs allow SARS-CoV-2 infection, but this can be reversed in part by dexamethasone
(A) Plots showing co-expression of spike binding domain with ACE2 on CD45+ CECs but not CD45− CECs from PBMCs from a COVID-19 patient.
(B) Image stream plots of spike protein interaction/binding with ACE2 on CECs in PBMCs from a COVID-19 patient; bright field (BF) plus spike staining (BF + Spike).
(C and D) (C) Cellular RNA level in CECs and (D) viral RNA copies in culture supernatants of CECs without (w/o) or with (w) magnetofection with SARS-CoV-2.
(E) Viral RNA copies in the culture supernatant of total CECs or monocytes of COVID-19 patients infected with SARS-CoV-2 and measured by qPCR 24 h after infection.
(F) Cellular RNA changes relative to the housekeeping gene (RNP) in cell pellet of CECs and monocytes of COVID-19 patients infected with SARS-CoV-2 and measured by qPCR 24 h after infection.
(G) Viral RNA copies in culture supernatants of total CECs of COVID-19 patients versus CECs of the placental tissues from healthy deliveries 24 h after infection with SARS-CoV-2.
(H) Cellular fold RNA changes relative to the housekeeping gene (RNP) in cell pellets of total CECs from COVID-19 patients or the placental tissues from healthy deliveries 24 h after infection with SARS-CoV-2.
(I and J) (I) Histogram plots and (J) cumulative data of the expression of ACE2 on total CECs and immune cells lineages of COVID-19 patients treated with or without dexamethasone (1 or 2 μg/mL) overnight.
(K) Viral RNA copies in culture supernatants of CECs either untreated or treated with dexamethasone (2 μg/mL) for 24 h before infection with SARS-CoV-2 as measured 24 h after infection.
(L) Cellular fold RNA changes relative to the housekeeping gene (RNP) in cell pellets of CECs either untreated or treated with dexamethasone (2 μg/mL) for 24 h before infection with SARS-CoV-2 as measured 24 h after infection. Each point represents data from cells of one patient, infection studies are representative of three independent experiments. Bar, mean ± 1 standard error.