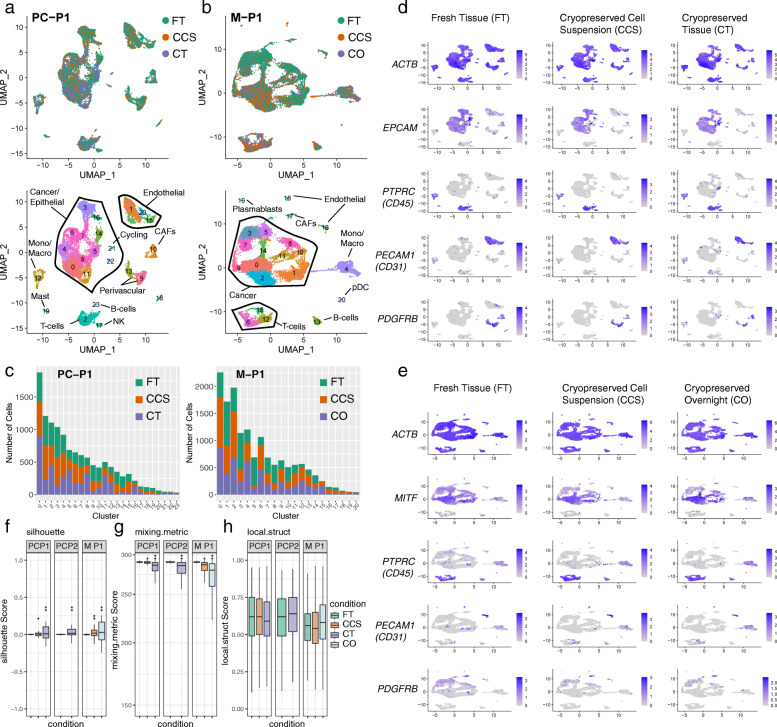
Fig. 2.
Cryopreservation allows for robust cell-type detection in clinical prostate cancer and melanoma samples. a UMAP visualisation of 18,331 cells sequenced across FT (green), CCS (orange) and CT (purple) from primary prostate cancer case PC-P1. UMAPs are coloured by cryopreserved replicate (top) and by cluster ID (bottom) with cell types annotations overlayed. Matched replicates were integrated using the Seurat v3 method. b UMAP visualisation as in a of 21,361 cells sequenced across FT (green), CCS (orange) and cryopreserved overnight (CO; purple) replicates from metastatic melanoma case M-P1. c Number of cells detected per cluster from PC-P1 and M-P1, highlighting the conservation of clusters detected in the FT samples following cryopreservation. Cells were downsampled to the lowest replicate size. d, e FeaturePlot visualisations of gene expression in prostate cancer (d) and melanoma (e) showing the conservation of the housekeeping gene ACTB and heterogeneous cancer/epithelial (EPCAM in d or MITF in e), immune (PTPRC/CD45), endothelial (PECAM1/CD31) and fibroblast/perivascular (PDGFRB) clusters following cryopreservation as FT, CCS and CT or CO. f–h Distribution of silhouette scores (f), mixing metric (g) and local structure metrics (h) of clustering following cryopreservation as analysed in Fig. 1e–g. Stars represent standard deviations: f silhouette scores s.d. 0.02–0.05* and s.d. > 0.05**; g mixing metrics s.d. 2–10* and s.d. > 10**; h local structure metrics s.d. > 0.05*