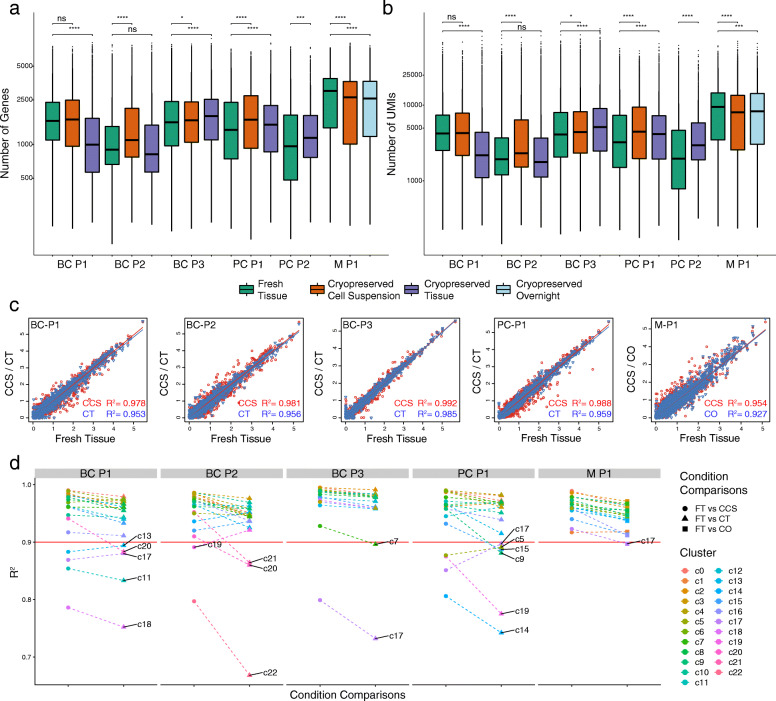
Fig. 3.
Cryopreservation maintains the integrity and complexity of single-cell transcriptomes in clinical human cancers. a, b Number of genes (a) and UMIs (b) detected per cell across all FT, CCS, CT and CO replicates from breast (BC-P1, BC-P2 and BC-P3), prostate (PC-P1 and PC-P2) and melanoma samples (M-P1). Sequencing libraries were downsampled to equal number of mapped reads per cell using the cellranger aggregate function to account for differences in sequencing depth. Note that only one CCS replicate in M-P1 (orange) and one CT replicate in BC-P1 (purple) had significantly lower number of genes and UMIs per cell compared to their matching FT replicate. Statistical significance was determined using an unpaired Student’s t test. P values denoted by asterisks: *p < 0.05, p < 0.01, *p < 0.001 and ****p < 0.0001. c Pseudobulk gene correlations between FT cells with CCS (red line) and CT or CO (blue line) replicates. Correlation values (adjusted-R2) were computed using linear regression in R to model the log-normalised gene expression values between two replicates. In all cases, CCS replicates had higher R2 values compared to CT and CO comparisons. d Cluster-level gene correlations between FT cells with CCS (circle), CT (triangle) and CO (square) replicates show similar trends with pseudobulk gene correlations. Dotted lines join corresponding clusters between different comparison methods. Plasmablasts (c18 in BC-P1 and c22 in BC-P2) were the only cell type identified in multiple cases to have significantly lower correlations