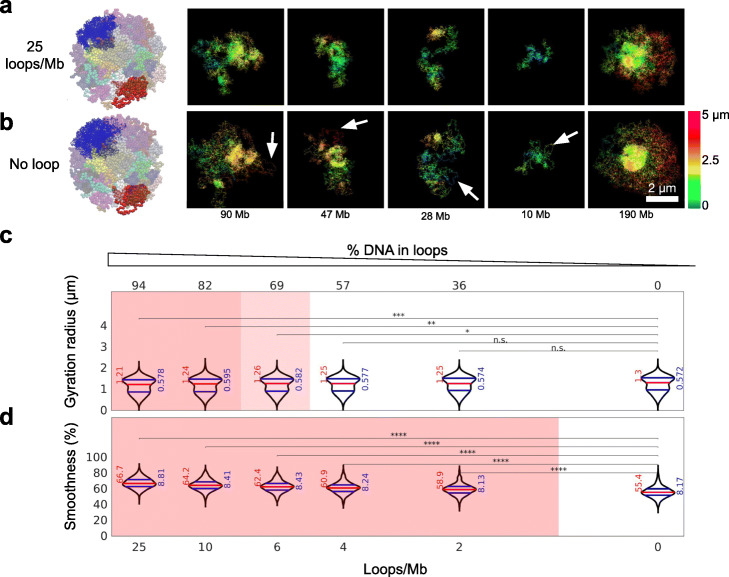
Fig. 5.
Modeling and quantifying the effect of cohesin-dependent loops on chromatin structure in silico. a,b Chromosome structures and images generated by our multi-scale polymer simulation. The five images show example 3D super-resolution views of simulated chromosome regions, with (a) or without (b) loops. The genomic size of each region is indicated and ranges from 10 to 190 Mb. A snapshot of the simulated model is shown to the left. For animated 3D views, see Additional file 5: Video S4. c. Violin plots show the distribution of gyration radii and smoothness as function of loop counts per Mb or, equivalently, percentage of DNA enclosed by loops. Pink shading indicates where median values differ with intermediate significance (p < 0.05, light shade), or with high significance (p < 0.01, darker shade) from values in absence of loops (two-sided rank sum tests)