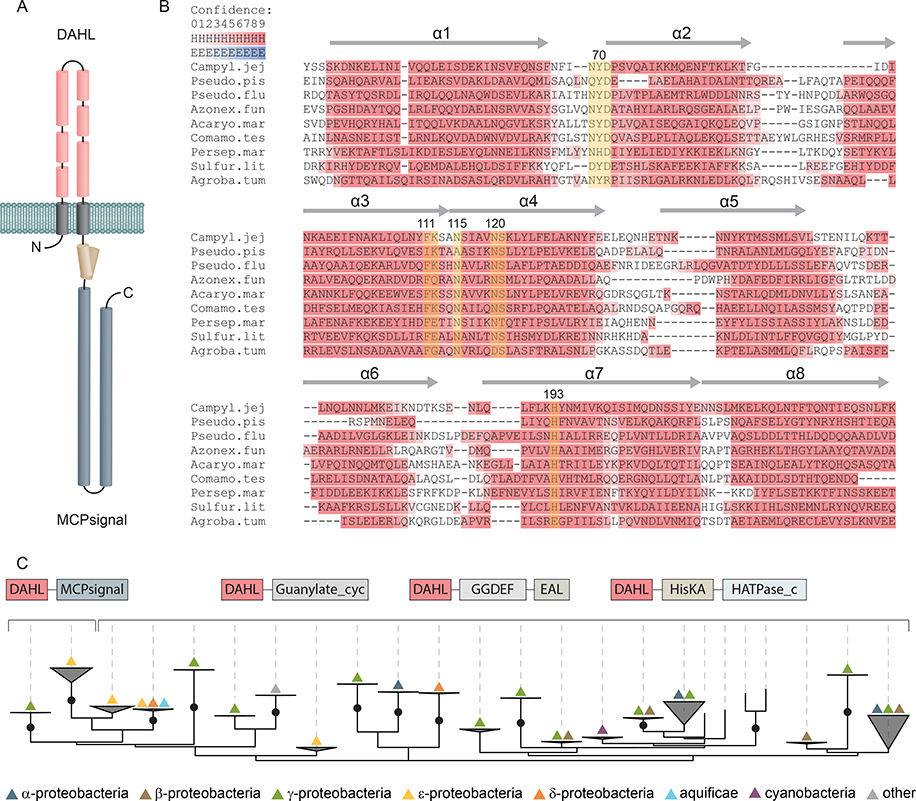
Fig. 1. Secondary structure prediction and phyletic distribution of the DAHL domain.
(A) Schematic representation of Tlp10 topology with the predicted architecture of the DAHL domain as the LBD, the transmembrane region, and the intracellular MCP signaling domain. (B) MSA and secondary structure prediction of periplasmic DAHL domains from various bacterial species. Predicted α-helical regions are labelled and highlighted in red. Conserved amino acid positions are highlighted in yellow. Representative sequences were selected from different clades on the phylogenetic tree. Campyl.jej: C. jejuni, Tlp10 WP_002865311.1; Pseudo.pis: Pseudoalteromonas piscicida, WP_099642651.1; Pseudo.flu: Pseudomonas fluorescens, WP_003172050.1; Azonex.fun: Azonexus fungiphilus, WP_121456889.1; Acaryo.mar: Acaryochloris marina, WP_012165516.1; Comamo.tes: Comamonas testosterone, WP_003076421.1; Persep.mar: Persephonella marina, WP_012676181.1; Sulfur.lit: Sulfurovum lithotrophicum, WP_052746078.1; Agroba.tum: A. tumefaciens, VirA WP_012478065.1. (C) Maximum likelihood phyletic distribution and protein architecture variations of DAHL domain–containing proteins with bootstrap support based on 500 replicates. Neighboring domains are indicated according to Pfam database nomenclature: MCPsignal, MCP signaling domain, PF00015; Guanylate_cyc, adenylate and guanylate cyclase catalytic domain, PF00211; GGDEF, diguanylate cyclase, PF00990; EAL, diguanylate phosphodiesterase, PF00563; HisKA, histidine kinaseA (phospho-acceptor) domain, PF00512; HATPase_c; -, DNA gyrase B-, phytochrome-like ATPases, PF02518. Bootstrap values more than 70% are indicated by black circles.