Fig. 1. Shikimatogenerans silvanidophilus, the symbiont of O. surinamensis.
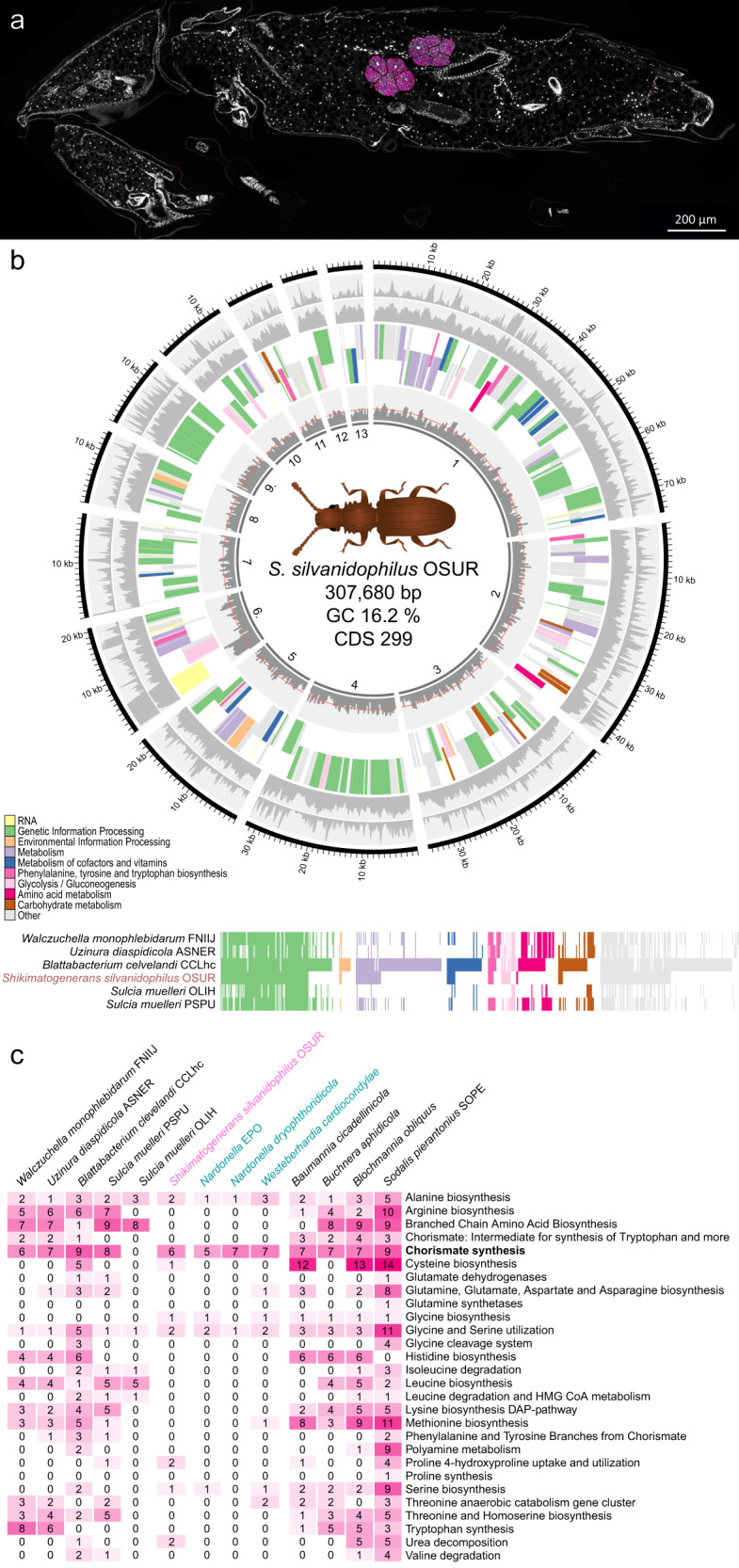
a Fluorescence in situ hybridization micrograph of a sagittal section of a 5-day-old O. surinamensis pupa stained with CFB563mod-Cy3 (magenta) and DAPI (white). b Circular representation of the draft genome of S. silvanidophilus. Single contigs are sorted clockwise by length. The outer gray circles denote coverage with long- and short-reads, respectively, the intermediate circles indicate annotated functional KEGG categories separated by direction of transcription (see legend for depicted categories). The inner gray circle denotes relative GC content and the average GC content of 16.2% by the red line. Below: comparison of the functional gene repertoires of Bacteroidetes symbionts in insects. Box colors are based on KEGG’s categories. c Detailled comparison of the amino acid metabolism gene repertoires between the S. silvanidophilus genome (pink), other Bacteroidetes symbionts (left) and Proteobacteria symbionts (right), some of which are known to exclusively provision tyrosine precursors to their insect host (blue).
