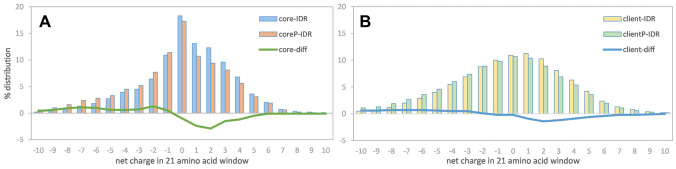
Figure 3.
Core (panel A) and client (panel B) net charge distributions, and phosphorylated counterparts (denoted P), are calculated from overlapping 21 amino acid windows within predicted IDRs. For each of core and client proteins, the difference (phosphorylated—unphosphorylated) is also shown, as a line plot. For simplicity, the net charge in each window is shown on the x-axis. In terms of net charge per amino acid, the displayed values range from a minimum of − 0.476 to a maximum of 0.476.