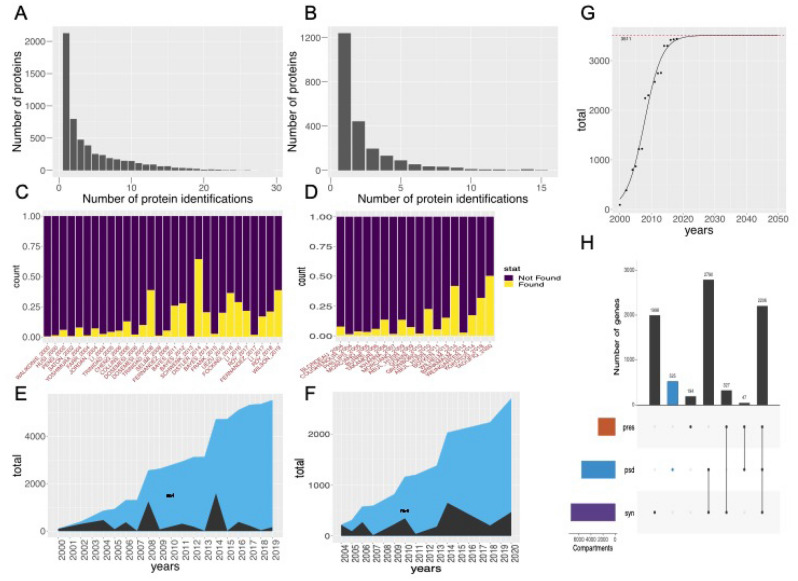
Figure 1.
(A) Discovery rate of new PSD proteins across 29 postsynaptic studies, where the number of proteins is plotted against the frequency of identification. 2091 PSP proteins have been observed just once. The most frequently found proteins (i.e. detected in 22, or more, studies out of the 29) include very well-known PSD proteins, for example: DLG4 (28/29), CAMK2A (27/29), INA (26/29), SPTBN1, CAMK2B, DLG2, NSF, GRIN2B, GRIN1 (25/29), BIAP2, BSN (24/29) (full list in Supplementary Table 2). (B) Discovery rate of new proteins analysed across 18 presynaptic studies. More than half of the proteins in the presynaptic proteome (1251) have been observed just once. The most frequent presynaptic genes include AP2B1, HSPA8, GNAO1, ACTB (15/17), STX1B, ATP6V0A1, STXBP1, ATP1A3, ATP6V1E1, SYT1, GNB1, TUBA1A, VAMP2, NSF, DNM1 (14/17) with full statistics available in Supplementary Table 3. (C) Contribution of each of 29 studies to the total number of PSP genes (purple—total number of genes, yellow—identified in this study). Two major jumps in the gross number of proteins identified occur in 2008, when 1249 new proteins were reported by14 and in 2014 with 2588 new proteins added by20. (D) Contribution of each of 18 studies to the total number of presynaptic genes (purple—total number of genes, yellow—identified in this study): two jumps in newly discovered proteins correspond to studies in years 2010 and 2014. (E) Accumulation of the new PSP genes (black) compared to the total datasets (blue) over years. (F) Accumulation of new presynaptic genes (black) compared to the total datasets (blue) over years. (G) Non-linear fit predicting the total size of “consensus” PSP (genes found in two and more studies, 3499) (P = 2.36E−11, residual standard error: 192.7 on 12 degrees of freedom) by year 2023 which, when compared to the current number (3438) indicates that our knowledge on PSP components, based on currently available methodologies, is close to saturation. (H) Overlap of three synaptic datasets: presynaptic, postsynaptic and synaptosomal. Bars correspond to the number of unique genes in each compartment and their intersections.