Abstract
For decades, more and more long non-coding RNAs (lncRNAs) have been confirmed to play important functions in key biological processes of different organisms. At present, most identified lncRNAs and those with known functional roles are from mammalian systems. However, lncRNAs have also been found in primitive eukaryotic fungi, and they have different functions in fungal development, metabolism, and pathogenicity. In this review, we highlight some recent researches on lncRNAs in the primitive eukaryotic fungi, particularly focusing on the identification of lncRNAs and their regulatory roles in diverse biological processes.
Keywords: lncRNAs, eukaryotic fungi, transcriptional interference, chromatin remodeling, RNA surveillance
Introduction
Recent researches on non-coding RNAs (ncRNAs) have expanded our knowledge of gene transcriptional regulation. These ncRNAs have been classified mainly into small ncRNAs and long non-coding RNAs (lncRNAs). Small ncRNAs are range from 17 nt to approximately 200 nt in length, while lncRNAs have lengths greater than 200 nt (Wilusz et al., 2009; Pasquinelli, 2012; Rinn and Chang, 2012; Fatica and Bozzoni, 2014). Small RNAs, including microRNAs, small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs), usually function as negative regulators that interfere with the expression of target RNAs, which to regulate diverse cellular, developmental, and physiological processes (He and Hannon, 2004; Bushati and Cohen, 2007; Carthew and Sontheimer, 2009; Ghildiyal and Zamore, 2009; Czech and Hannon, 2011). In comparison with small ncRNAs, lncRNAs have more complicated and flexible regulatory functions in numerous biological processes, including dosage compensation, alternative splicing, genomic imprinting, X-chromosome inactivation, and so on (reviewed in Wilusz et al., 2009; Hung and Chang, 2010; Lee, 2012; Cheetham et al., 2013; Kung et al., 2013; Fatica and Bozzoni, 2014). Like mRNAs, many lncRNAs are 5’-capped and 3’-polyadenylated. They are usually spliced products of RNA polymerase II (RNAPII). lncRNAs have been classified as sense, antisense, bidirectional, intronic, and intergenic based on their locations relative to adjacent encoding genes (Mishra and Kanduri, 2019). Regulatory lncRNAs are often expressed at specific development stages or in response to conditional changes of nutrition or environment (reviewed in Hung and Chang, 2010; Guttman and Rinn, 2012; Lee, 2012; Rinn and Chang, 2012). Also, some lncRNAs are translated into stable functional micropeptides, or function as a sponge to recruit microRNAs (Anderson et al., 2015; Paraskevopoulou and Hatzigeorgiou, 2016; Yeasmin et al., 2018; Zhao et al., 2020). Some lncRNAs form circlular RNAs (circRNAs) and act as transcript effectors that regulate—the target gene expression (Cesana et al., 2011; Sanchez-Mejias and Tay, 2015; Thomson and Dinger, 2016; Zhong et al., 2018). All these researches have enhanced our understanding about the functions of non-coding RNAs in different kinds of biological processes.
At present, a lot of lncRNAs such as H19, Xist, MALAT1, and HOTAIR have been well characterized in mammals through genetic and molecular studies (Brown et al., 1991; Tripathi et al., 2010; Hung et al., 2011; Nagano and Fraser, 2011). In plants, since the first plant lncRNA Enod40 was discovered in 1994 (Crespi et al., 1994), several plant lncRNAs have also been functional identified (Franco-Zorrilla et al., 2007; Swiezewski et al., 2009; Ariel et al., 2014, 2020; Kim and Sung, 2017; Wu et al., 2020). These lncRNAs from mammals and plants can regulate gene expression at epigenetic, transcription, and post-transcription levels and widely take part in various physiological and pathological processes (reviewed in Rinn and Chang, 2012; Cheetham et al., 2013; Fatica and Bozzoni, 2014). However, research on the mechanisms of lncRNAs functions in eukaryotic microbes is still in its infancy. Although 80% of budding and fission yeast genomes are transcribed and most of the transcripts are translated into proteins, many non-coding transcripts still exist (Christie et al., 2004; David et al., 2006; Nagalakshmi et al., 2008; Jacquier, 2009). With the development of advanced biotechnologies, such as high-resolution tilling arrays and high-throughout sequencing (e.g., RNA-Seq), numerous lncRNAs have also been found in several fungi. Although the molecular functions of most fungal ncRNAs remaining elusive, a small number of functions in model organisms, such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Neurospora crassa, have been determined. These fungal lncRNAs have different functions in a wide variety of biological processes, including fungal development, mating-type regulation, metabolism, cell differentiation, sporulation, and nutrient metabolism (Donaldson and Saville, 2012). Because of the vital roles of fungi in ecosystems, studying their lncRNAs may help us expand our knowledge of ncRNA-related mechanisms in different species. In this review, we provide a recent snapshot of lncRNAs in eukaryotic microbes. We aim to describe the regulatory functions of fungal lncRNAs in gene regulation and development.
Identification and Classification of lncRNAs in Fungi
Advancements in biotechnologies have led to the identification of non-coding transcripts in fungi. Initially, in 2006, tiling arrays were used to identify the lncRNAs of the S. cerevisiae genome (David et al., 2006; Samanta et al., 2006). However, this method produced a high level of background noise and was difficult to interpret with a high degree of confidence. Thus, with the advancement of next-generation sequencing, strand-specific RNA sequencing has been successfully used to identify lncRNAs in different fungal organisms, including S. cerevisiae, S. pombe, and N. crassa. At present, the RNA-seq technique is the most powerful method to identify fungal lncRNAs because it reveals both already known sequences and novel variants (Piskol et al., 2013). Furthermore, lncRNA microarrays, which is different from conventional mRNA sequence-based gene expression microarrays, have been developed to assess the biological relevance of lncRNAs in pathological conditions (Huang et al., 2018). Using high-throughput sequencing and bioinformatics methods, putative ncRNAs are obtained. Moreover, several biological molecular methods have provided powerful platforms for detecting the transcriptional regulatory functions of lncRNAs (McDonel and Guttman, 2019). For example, chromatin immunoprecipitation (ChIP) and related derivative technologies, such as RIP (RNA immunoprecipitation), CLIP (UV crosslinking and immunoprecipitation), ChIRP (chromatin isolation by RNA purification), CHART (capture hybridization analysis of RNA targets), and RAP (RNA antisense purification), have been used to investigate the actions of lncRNAs that interact with RNA, DNA, proteins, and nucleic acids (Cao et al., 2019). Additionally, techniques such as genetic modification, overexpression or knockdown strategies, and phenotype analyses have been used to determine the functional importance of lncRNAs in different species.
In general, lncRNAs are present at relatively low abundance levels and are not effectively detected using empirical techniques because they likely exist in a dynamic equilibrium that is constantly monitored by the RNA surveillance machinery (Nair et al., 2020). The RNA surveillance system monitors each step of RNA biogenesis, including 5′-capping, 3′-splicing end formation, polyadenylation, nuclear export, and full-length protein translation (Pefanis et al., 2015; Nair et al., 2020). In yeast, RNA surveillance-mediated RNA degradation is mainly classified as nuclear and cytoplasmic degradation. In the nucleus, the RNA exosome complex has critical functions in the 3′–5′ RNA degradation associated with early transcription termination (Houseley et al., 2006; Schmid and Jensen, 2008; Fraga de Andrade et al., 2020). The eukaryotic exosome complex includes nine subunits: six distinct proteins form as a “ring” and three RNA-binding proteins form a “cap.” Interestingly, two additional subunits, Dis3 (Rrp44) and Rrp6 (Exosc10), provide the enzymatic activity of the exosome complex (Houseley et al., 2006; Januszyk and Lima, 2010; Wasmuth and Lima, 2012). Rrp6 is a nuclear-specific 3′–5′ distributive exoribonuclease (Lykke-Andersen et al., 2009; Wasmuth et al., 2014). In addition, efficient RNA degradation by the exosome requires the polyadenylation of the Trf4-Air2-Mtr4p (TRAMP) complex (Houseley et al., 2006). Although the eukaryotic RNA exosome complex functions in both the nucleus and the cytoplasm, studies have found that the transcripts in the latter were mainly degraded by the 5′–3′ exonuclease Xrn1 (Nagarajan et al., 2013). Before the Xrn1-mediated degradation, Dcp2 needs to remove the 5′-caps (Garneau et al., 2007).
Eliminating key components of the RNA surveillance system has enabled researchers to identify some previously undescribed lncRNAs in cells. Thus, a series of novel lncRNAs has been identified in fungi. For example, 925 transcripts encoded in intergenic regions accumulate in S. cerevisiae mutants lacking the exosome subunit Rrp6, while limited expression changes have been identified in most of the open reading frame transcripts in both wild-type and Rrp6 mutant (Wyers et al., 2005). These identified non-coding transcripts are transcribed by RNAPII, capped, and polyadenylated. More importantly, they lack promoter elements and the rapid degradation by nuclear exosomes found in wild-type cells. Consequently, these ncRNAs are referred to as cryptic unstable transcripts (CUTs) (Table 1; Wyers et al., 2005; Berretta et al., 2008). The high-resolution genomic map of yeast revealed 1,496 CUTs, and numerous new CUTs may be associated with regulatory mechanisms (Neil et al., 2009). Another full inactivation of the exosome resulted in at least 1,600 CUTs (Gudipati et al., 2012). Interestingly, aberrant polyadenylated transcripts from small nucleolar RNA genes also accumulate in Rrp6 mutants (Davis and Ares, 2006). After deleting the decapping enzyme Dcp2, more than 100 lncRNAs accumulate in S. cerevisiae, indicating that decapping also control the stability of lncRNAs (Ramaiah et al., 2012). Also, a kind of cryptic non-coding transcripts named Xrn1-sensitive unstable transcripts (XUTs) have been observed with the absence of Xrn1 in budding yeast cells (Table 1). XUTs can modulate the chromatin structure of the promoter regions to repress the expression of sense transcripts (van Dijk et al., 2011). It is interesting that both the Dcp2-sensitive lncRNAs and XUTs are often antisense to encoding genes.
TABLE 1.
Classification of lncRNAs in fungi.
| Name | Related enzyme or protein | How to found | Functions | References |
| CUTs | Rrp6 (3′–5′ distributive exoribonuclease) | With the inactivation of Rrp6 | Nuclear, regulatory mechanisms | Wyers et al., 2005; Gudipati et al., 2012 |
| Dcp2-sensitive lncRNAs | Dcp2 (decapping enzyme of 5′-caps) | With the inactivation of Dcp2 | Cytoplasm, function unknown | Ramaiah et al., 2012 |
| XUTs | Xrn1 (5′–3′exonuclease) | With the inactivation of Xrn1 | Cytoplasm, modulate the chromatin structure of the promoter regions to repress the expression of sense transcript | van Dijk et al., 2011 |
| SUTs | May be substrates of Xrn1 | Expressed in vegetative yeast cells | Regulate vegetative growth | Xu et al., 2009 |
| MUTs | Most degraded by the exosome component Rrp6 | Expressed during Mitotic growth | Regulate the sense genes during meiosis | Lardenois et al., 2011 |
| TERRAs | Rat1(5′-end exonuclease) | Degraded by Rat1 | Control telomere length | Luke et al., 2008 |
| CD-CUTs | Cytoplasmic non-sense-mediated mRNA decay pathway | With the inactivation of cytoplasmic non-sense-mediated mRNA decay pathway | Interferes with the binding of RNAPII and transcriptional activators by repressing bona fide promoters under repressive conditions. | Toesca et al., 2011 |
| NUTs | Nrd1 (RNA-binding factor) | With the mutant of Nrd1 | Sensitive to the early Termination of lncRNA transcription | Schulz et al., 2013 |
| Set2-repressed antisense transcripts | Set2 (histone methyltransferase) | With the deletion of Set2 | Undetermined | Venkatesh et al., 2016 |
Beside the identification of those non-coding RNAs degraded by the RNA surveillance system, a new set of transcripts, named stable unannotated transcripts (SUTs), which are expressed with low abundance in vegetative yeast cells, have also been identified (Table 1; Xu et al., 2009). Interestingly, most SUTs may be substrates of Xrn1 because they accumulated in an Xrn1 mutant grown in a YPD (Yeast Extract Peptone Dextrose) medium (Xu et al., 2009). Furthermore, non-coding transcripts have not only been revealed during vegetative growth but also during mitotic growth. Meiotic unannotated transcripts (MUTs), which accumulate during mitotic growth in S. cerevisiae, have also been discovered recently (Table 1; Lardenois et al., 2011). Some MUTs are transcribed on the antisense strands of coding genes, implying that they may regulate the sense genes during meiosis. Additionally, most MUTs are degraded by the exosome component Rrp6, suggesting that they are a subclass of CUTs (Lardenois et al., 2011).
Interestingly, a pervasive yeast telomeric repeat-associated cryptic ncRNAs (TERRAs), which is degraded by the nuclear 5′-end exonuclease Rat1, has also been found to control telomere length (Table 1; Luke et al., 2008). Recently, a group of cryptic transcripts, named cytoplasmically degraded-CUTs (CD-CUTs), have been found in with the inactivation of the cytoplasmic non-sense-mediated mRNA decay pathway (Table 1; Toesca et al., 2011). The transcription of these CD-CUTs interferes with the binding of RNAPII and transcriptional activators by repressing bona fide promoters under repressive conditions.
Thus, identifying ncRNAs involves surveying stabilized or enriched RNAs in mutants defective in RNA degradation pathway provides novel information on the genome-wide occupancy of the transcriptional machinery. In addition, a class of ncRNAs termed Nrd1-unterminated transcripts (NUTs), which is sensitive to the early termination of lncRNA transcription, has also been identified in the RNA-binding factor Nrd1 mutant cells (Table 1; Schulz et al., 2013). Similarly, Set2-repressed antisense transcripts have been identified using the deletion of the histone methyltransferase Set2 (Table 1; Venkatesh et al., 2016). However, although it still not clear whether these unstable transcripts form explicit mechanisms or represent an unexpected side effect of transcriptome surveillance, these types of unstable transcripts result in the silencing of their target genes. Thus, they are usually used to control pervasive transcription and offer protection from gene silencing throughout the life cycle.
Regulatory Mechanisms of lncRNAs in Fungi
In general, functional lncRNAs can bind with DNA, RNA, or protein to regulate the expression of target genes through transcriptional interference, promoter occlusion, and/or recruit epigenetic chromatin-modifier recruitment via cis- or trans- model (Mishra and Kanduri, 2019). Because S. cerevisiae lacks apparent homologs of Argonaut or Piwi-like proteins, those complexes, such as RNA-induced silencing complex (RISC) and RNA-induced transcriptional silencing (RITS) which mediate the interactions between regulatory siRNAs and their nucleotide targets, have not been observed in this species (Aravind et al., 2000; Drinnenberg et al., 2009, 2011). However, S. cerevisiae may have evolved novel post-transcriptional regulatory strategies to adapt to the loss of RNAi. For example, it can utilize the sequence-specific Ty1 retrotransposon to regulate the ncRNA-mRNA/DNA interactions (Berretta et al., 2008). In S. pombe, siRNAs are incorporated into RNA-induced transcriptional silencing complexes. This complex then recruit the histone methyltransferase Clr4 and the S. pombe homolog of metazoan heterochromatin protein Swi6 to promote the transcriptionally silent heterochromatin formation (Moazed et al., 2006; Zhang et al., 2011). Moreover, a novel RNAi-independent cosuppression of long terminal repeat-retroelements have also been found in the phytopathogenic fungus Magnaporthe oryzae (Murata et al., 2007). In addition, lncRNAs such as meiRNAs, TERRAs, and telomerase RNAs can act as scaffolds or attract proteins to alter their functions in yeast (Ding et al., 2012; Cusanelli and Chartrand, 2015). They are presented in detail in the following sections.
Depending on their modes of gene regulation, lncRNAs may also be categorized as cis- or trans-acting (Camblong et al., 2007, 2009). The trans-acting lncRNAs often regulate target genes through pre-translational and/or post-translational mechanisms and act as guides or scaffolds for chromatin-remodeling complexes (Guil and Esteller, 2012). However, most of the functional lncRNAs identified in fungi mainly perform their functions in cis using different regulatory strategies. At present, the simplest and probably most common regulatory strategy that has been reported among the fungal lncRNAs is transcriptional interference, especially interfering with the transcription of proximally located genes. lncRNAs govern the expression of their adjacent genes, both sense and antisense, in a repressive or activating manner. Also, their regulatory mechanisms are varied, including blocking the transcription machinery, modulating the nucleosomal arrangement, and stimulating regulatory factors binding or dissociation. There are numerous examples of lncRNAs in fungi exerting transcriptional interference, such as nc-tgp1, prt, and prt2 (reviewed in Kornienko et al., 2013; Vance and Ponting, 2014; Shuman, 2020). Moreover, numerous functionally identified lncRNAs interact physically with RNAPII complexes to regulate mRNAs transcription (Goodrich and Kugel, 2006).
Another regulatory strategy of lncRNAs is chromatin remodeling. They have been identified as the regulators of chromatin structure because their transcription recruits various histone/DNA modification enzymes, such as methylation of histone 3 (H3K4me3, H3K9me3, H3K27me3, and H3K36me3), to alter chromatin structure and modify histones, which influences the recruitment or activity of transcription factors (Martens et al., 2004, 2005; Hongay et al., 2006; Uhler et al., 2007; Houseley et al., 2008). Chromatin remodeling guided by lncRNAs contributes mechanistically to the establishment of chromatin structure and the maintenance of epigenetic memory. Antisense transcript-mediated chromatin remodeling may occur in cis or trans. For instance, the antisense transcript Ty1AS inhibits its retro-transposition in S. cerevisiae in trans (Berretta et al., 2008). All the metabolic stress-inducing lncRNA mlonRNA from S. pombe and ncASP3 and the antisense lncRNA of the CDC28 gene in S. cerevisiae are involved in stress responses through affecting the chromatin organization (Nadal-Ribelles et al., 2014).
Functional Diversity of lncRNAs in Fungi
Although numerous long non-coding transcripts have been identified in fungi, only a few have been functionally identified (Table 2). However, these functional studies showed that lncRNAs in fungi are involved in complex regulatory networks, performing essential cellular tasks, including the regulation of meiosis and mating, metabolisms, cell aging, circadian rhythms, and pathogenesis (Table 2). Here, we describe some functionally identified lncRNAs in fungi to provide an understanding of their specific regulatory roles.
TABLE 2.
Experimentally characterised lncRNAs in fungi.
| Species | Non-coding | Gene | Antisense/sense | Cis/trans | Coactivator | Cellular | Regulatory | References |
| RNA name | name | to mRNA | regulation | process | mechanism | |||
| S. cerevisiae | GAL10-ncRNA | GAL10/GAL1 | Antisense/Sense | cis | reb1 | Galactose utilization | Histone modification | Houseley et al., 2008 |
| S. cerevisiae | GAL4 lncRNA | GAL4 | Antisensense | cis | /* | Galactose utilization | / | Geisler et al., 2012 |
| S. cerevisiae | ncASP3 | ASP3 | Sense | cis | / | Nitrogen starvation | Histone modification, chromatin remodeling | Huang et al., 2010 |
| S. cerevisiae | PHO84 antisense transcripts | PHO84 | Antisense | trans, cis | Hda1/2/3 | Phosphate metabolism, Cell aging | Histone modification | Camblong et al., 2007 |
| S. cerevisiae | REM2 | IME4 | Antisense | cis | / | Sexual differentiation and mating-type control | Transcriptional interference | Hongay et al., 2006 |
| S. cerevisiae | IRT1 | IME1 | Sense | cis | Rem1 | Sexual differentiation and mating-type control | Histone modification, transcriptional interference | van Werven et al., 2012 |
| S. cerevisiae | REM3 | ZIP2 | Antisense | cis | / | Sexual differentiation and mating-type control | Transcription interference | Gelfand et al., 2011 |
| S. cerevisiae | SRG1 | SER3 | Sense | cis | SAGA and Swi/Snf, Cha4, | Serine synthesis | Transcriptional interference | Martens et al., 2005 |
| S. cerevisiae | pHO-lncRNA | HO | Sense | cis | / | Mating type interconversion, cell-cycle | Transcriptional interference, nocleosome repositioning | Yu et al., 2016 |
| S. cerevisiae | PWR1 | FLO11 | Antisense | cis | / | Cell-cell adhesion | Transcriptional interference | Bumgarner et al., 2009 |
| S. cerevisiae | ICR1 | FLO11 | Sense | cis | / | Cell-cell adhesion | Promoter occlusion, silencing | Bumgarner et al., 2009 |
| S. cerevisiae | AS-PHO5 | PHO5 | Antisense | cis | / | Phosphate metabolism | Activate PHO5 transcription during phosphate starvation | Uhler et al., 2007 |
| S. cerevisiae | Antisense lncRNA of CDC28 | CDC28 | Antisense | cis | / | Osmostress | Chromatin remodeling | Nadal-Ribelles et al., 2014 |
| S. cerevisiae | Antisense lncRNA of Ty1 | Ty1 | Antisense | trans | / | Transponson silencing | RNA-interference | Berretta et al., 2008 |
| S. cerevisiae | TERRA | Telomerase RNA | / | / | / | Telomere replication | Regulation of telomerase activity, heterochromatin formation | Luke et al., 2008 |
| S. cerevisiae | TLC1 | Telomerase RNA | / | / | Est1, Est2, and Est3 | Telomere replication | Scaffold for telomerase complex | Gallardo et al., 2008 |
| S. cerevisiae | ADF1 | MDF1 | Antisense | cis | / | Vegetative growth | Transcriptional suppression | Li et al., 2010 |
| S. pombe | prt/nc-pho1 | pho1 | Sense | cis | Pho7 | Phosphate metabolism. | Chromatin remodeling | Chatterjee et al., 2016 |
| S. pombe | nc-tgp1 | tgp1 | Sense | cis | Pho7 | Phosphate metabolism | Transcriptional interference | Ard et al., 2014 |
| S. pombe | prt2 | pho84(prt/pho1) | Sense | cis | / | Phosphate metabolism | Transcriptional interference | Garg et al., 2018 |
| S. pombe | mlonRNAs | fbp1 | Sense | cis | United States1, United States2 | Glucose starvation | Chromatin remodeling | Hirota et al., 2008 |
| S. pombe | SPNCRNA .1164 | atf1 | / | trans | / | Oxidative stress | Activation | Leong et al., 2014 |
| S. pombe | meiRNA-S and L | sme2 | Sense | cis | Mei2, Mmi1 | Meiosis | Decoy of Mmi1 | Shichino et al., 2014 |
| S. pombe | TER1 | telomerase RNA | / | / | / | Telomere replication | Scaffold for telomerase complex | Leonardi et al., 2008 |
| T.reesei | HAX1 | cellulase genes | / | trans | Xyr1 | Cellulose metabolism | Activation | Till et al., 2018 |
| C. heterost rophus. | Antisense of tramscription factor CMR1 | Melanin gene cluster | Antisense | trans | MAPK pathway | Melanin biosynthesis | Regulate the transition of the melanin gene cluster | Eliahu et al., 2007 |
| N. crassa | qrf | frq | Antisense | cis | / | Rhythmic conidiation | Chromatin modifications and the premature termination of transcription | Kramer et al., 2003 |
| U. maydis | Antisense to gene um02151 | um02151 | Antisense | / | / | Pathogenesis | Unknown | Donaldson and Saville, 2013 |
| C. neoformans | RZE1 | Znf2 | / | / | / | Yeast-to-hypha transition | Unknown | Chacko et al., 2015 |
| F. oxysporum | Fo-carP | carS | / | / | / | Carotenoid biosynthesis | Active expression of the carotenoid genes | Parra-Rivero et al., 2020 |
| F. fujikuroi | Ff-carP | / | / | / |
*Details unknown.
Cell Cycle or Meiosis Control
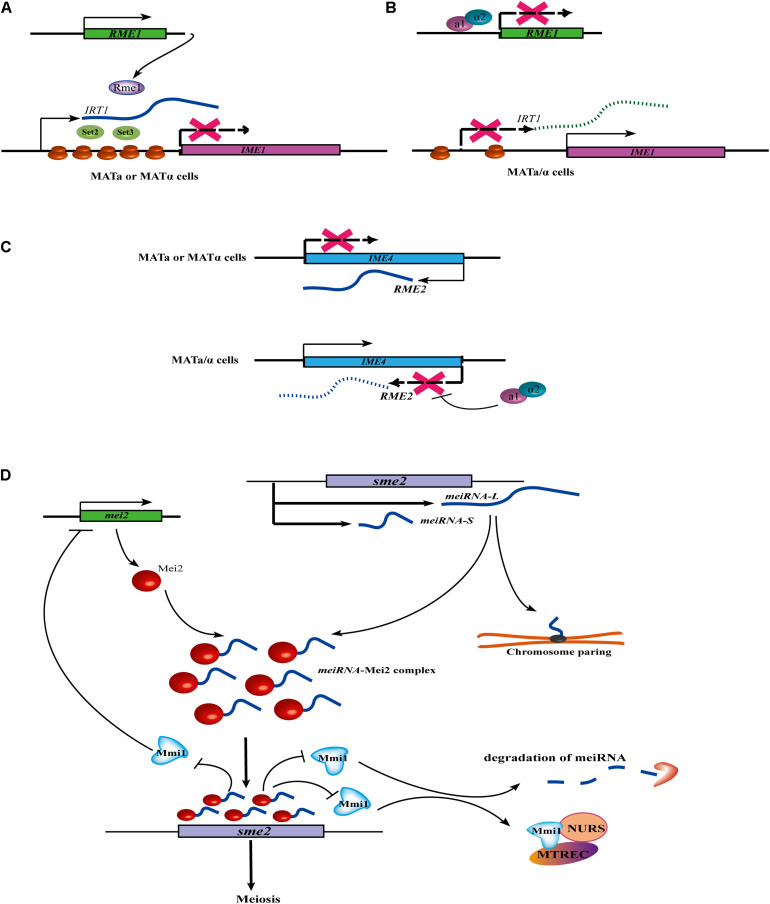
In yeast, the mating of haploid cells with the opposite mating type (MATa and MATα) can produces MAT a/α diploid cells. In S. cerevisiae, the induction of meiosis and sporulation are dependent on the transcriptional activation of the IME1 (Inducer of Meiosis 1) gene. At present, at least three lncRNAs (IRT1, RME2, and RME3) have been identified to control the switch of mating-type in S. cerevisiae. IRT1 arises from the same strand as the IME1 promoter. A meiosis-repressive transcription activator, Rme1, acts as a coactivator to induce the production of the lncRNA IRT1. Then, IRT1 recruits the histone methyltransferase Set2 and the histone deacetylase Set3 to inhibit IME1 expression through establishing repressive chromatin at the IME1 promoter (Figure 1A; van Werven et al., 2012). Further evidence has revealed that the a1/α2 heterodimer inhibits the expression of RME1. Without the repression of IRT1, IME1 is successfully transcribed (Figure 1B; van Werven et al., 2012). RME2 is another lncRNA that inhibits the transcriptional elongation of a putative RNA methyltransferase, IME4, to prevent germ cell differentiation in MATa or MATα haploid cells. However, in MATa/α diploid cells, the a1/α2 heterodimer binds at a conserved site located downstream of the IME4, leading to the repression of RME2 and the induction of IME4 (Figure 1C; Hongay et al., 2006). Moreover, RME3 represses the expression of its adjacent gene, ZIP2, which is required for chromosomal pairing during meiosis (Gelfand et al., 2011).
FIGURE 1.
Cell cycle or meiosis control of lncRNA in fungi. (A) In MATa or MATα haploid cells, the expression of IRT1 is induced by a transcription activator Rme1. Then transcribed IRT1 can recruit the histone methyltransferase Set2 and the histone deacetylase Set3 at the IME1 promoter, finally leading to the inhibit of IME1 expression. (B) In the homozygous MATa/α diploid cells, the a1/α2 heterodimer can inhibit the expression of REM1, so IRT1 can’t be expressed without the induction of Rme1. IME1 can successfully transcript without the repression of IRT1. (C) In MATa or MATα haploid cells, lncRNA REM2 inhibits transcription elongation of IME4 to prevent germ cell differentiation. However, in MATa/α diploid cells, the a1/α2 heterodimer represses the transcription of REM2 by binding at a conserved site located downstream of the IME4, thus allowing for induction of IME4. (D) meiRNA regulates the expression of meiotic gene and chromosome pairing. In meiotic cells, two isoforms of the meiRNA (i.e., meiRNA-S and meiRNA-L) are transcribed and form a meiRNA-Mei2 complex by interacting with RNA-binding protein Mei2. The Mei2-meiRNA complex blocks the RNA-binding protein Mmi1, thus allowing the sme2 to be translated, thus initiating meiosis. During meiotic prophase in S. pombe, the ends of each chromosome are tied up to the spindle pole body. At the sme2 loci on chromosome II, the formation of a robustly chromosome pairing is dependent on meiRNA. Interestingly, Mmi1 also inhibits Mei2 by forming a double negative feedback loop. In addition, Mmi1 can interact with a nuclear complex called Meiotic gene silencing complex MTREC/NURS MTREC (MTl1-REd1 Core) or NURS (NUclear RNA Silencing) in meiotic transcript elimination.
Additionally, the pHO-lncRNA, which originates at ∼2,700 bp upstream of the HO gene, is responsible for mating-type interconversion during cell-cycle re-entry after a pheromone-dependent arrest in G1. It is induced to force nucleosome repositioning at the locus of the downstream located HO endonuclease in S. cerevisiae (Yu et al., 2016). The production of a pheromone (i.e., the α-factor) and nucleosome rearrangement is induced with the transcription of pHO-lncRNA, leading to the activating signal loss of the Swi4/Swi6 cell-cycle box-binding factor from the HO promoter. The displacement of the binding factor prevents the HO expression, which blocks mating-type interconversion during re-entry into the cell cycle (Yu et al., 2016).
Interestingly, in contrast to S. cerevisiae, the meiosis mechanism in S. pombe is controlled by a lncRNA termed meiRNA with a different regulatory strategy. meiRNA is not involved in the induction of meiosis, but in meiotic progression and also chromosomal pairing (Ding et al., 2012). In meiotic cells, two isoforms of the meiRNA (meiRNA-S and meiRNA-L) are transcribed from the locus and physically interact with RNA-binding protein Mei2 to form a meiRNA-Mei2 complex (Figure 1D). In the nucleus, the Mei2-meiRNA complex blocks another RNA-binding protein, Mmi1, a crucial inhibitor of meiosis, thus allowing sme2 to escape degradation and be translated in the cytoplasm, initiating meiosis. Moreover, a robustly meiRNA-dependent chromosome pairing at the sme2 loci was also discovered (Figure 2). Interestingly, Mmi1 also inhibits Mei2 by forming a double-negative feedback loop (Shichino et al., 2014). In addition, Mmi1 interacts with a nuclear complex called Meiotic gene silencing complex MTREC/NURS MTREC (MTl1-REd1 Core) or NURS (NUclear RNA Silencing) that is involved in meiotic transcript elimination (Shichino et al., 2020).
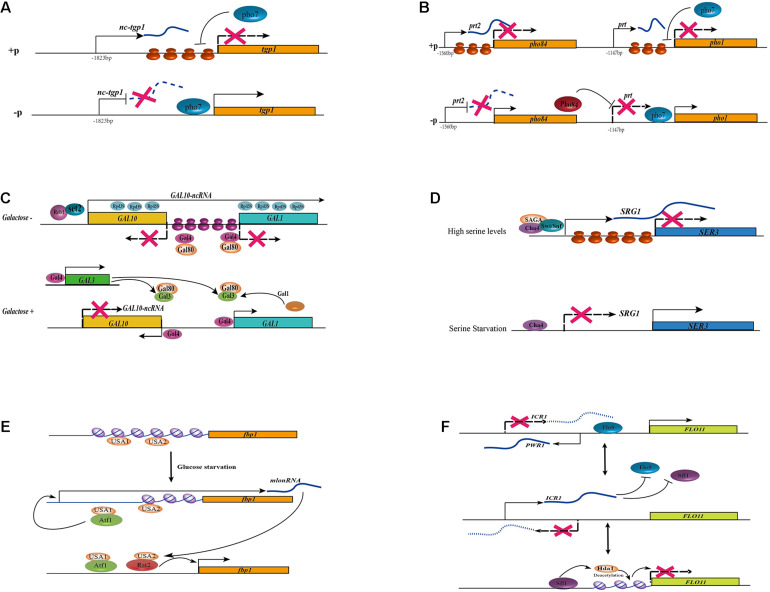
FIGURE 2.
Different regulatory mechanisms of lncRNAs in fungi. (A) In the addition of phosphate, the lncRNA nc-tgp1 initiates at 1,823 nt upstream of the tgp1 mRNA start site. The nucleosome density at the tgp1 promoter is increased with the transcription of nc-tgp1, which leads to the displacement of Pho7 from the tgp1 promoters, then results in the derepression of tgp1. The transcription of nc-tgp1 is reduced during phosphate deprivation, leading to a decrease in the nucleosome occupancy and expression of tgp1. (B) In the addition of phosphate, the lncRNA prt2 arises from the 1,560 nt upstream of the pho84 mRNA transcription start site. The transcription of lncRNA prt2 blocks the expression of its sense gene pho84. Similarly, the transcription of lncRNA prt blocks the expression of its sense gene pho1 by increasing nucleosome density at the pho1 promoter and displacing Pho7 from the pho1 promoters, leading to the derepression of pho1. The expression of lncRNA prt2 is depressed during phosphate starvation, leading to the production of Pho84, which acts as a repressor of prt, finally resulting in the expression of pho1. (C) lncRNA GAL10-ncRNA regulate the expression of GAL genes. In the absence of galactose, an antisense transcript GAL10-ncRNA that was initiated near the 3’end of GAL10 and terminated in the GAL1 coding region. Transcription of this antisense transcript is dependent on the transcriptional activator Reb1 and Set2 methyltransferase and histone deacetylation activities in cis, thus resulting in silencing of the whole GAL locus. In the absence of galactose, the activity of Gal4 activator is suppressed by Gal80. In the presence of galactose, Gal4 is released, leading to transcriptional activation of the GAL gene by sequestering Gal80 with Gal. Both GAL3 and GAL1 genes can be activated by Gal4 activator, thus forming two positive feedback loops and leading to stable Gal4 release. (D) The transcription of lncRNA SRG1 represses the adjacent SER3 gene under serine-rich conditions. When serine is available to the cells, SRG1 transcription is turned on via a serine-dependent activator Cha4, a coactivator complex SAGA and the ATPase subunit of the Swi/Snf chromatin remodeling complex, leading to the increase in nucleosome occupancy at the SER3 promoter, then repress the expression of SER3. In the absence of serine, although the Cha4 constitutively bound to the promoter of lncRNA SRG1, its transcription is repressed and the expression of SER3 is activated. (E) lncRNA mlonRNA regulates fbp1 genes through chromatin remodeling. There are two cis-acting regulatory elements in the upstream region of fbp1: upstream activating sequence1 (UAS1) and (UAS2). During glucose starvation, UAS1 can recruit the CREB/ATF-family transcription factor Atf1, then induce the expression of non-coding transcripts upstream of the fbp1 gene, the so-called mlonRNAs. These metabolic stress-induced lncRNAs trigger the disruption of the chromatin structure around UAS2, which in turn allows for binding of the zinc-finger transcription factor Rst2 to UAS2. The chromatin around the TATA box of the fbp1 gene is then modified and the stepwise chromatin remodeling through the fbp1 promoter results in and the eventual full expression of fbp1 in the absence of glucose. (F) The expression of the FLO11 gene is indirectly controlled by two lncRNAs, ICR1 and PWR1. The lncRNA ICR1 (3.2 knt) is transcribed in the same direction as FLO11 from the upstream intergenic region while PWR1 (1.2 knt) is antisense to ICR1. Both Flo8 and Sfl1 compete for binding to the FLO11 promoter, and also determining which of the two lncRNAs is expressed. histone deacetylase Hda1-mediated deacetylation condenses the chromatin at the Flo8 and Sfl1 binding sites. When Flo8 binding over Sfl1, the transcription of PWR1 is activated then PWR1 interferes with ICR1 via transcriptional interference, leading to the expression of FLO11. Conversely, the binding of Sfl1 recruits histone deacetylase Hda1 and PWR1 is not transcribed, enabling ICR1 transcription to interfere with FLO11.
Metabolisms and Nutrition
Phosphate Metabolisms
The fission yeast S. pombe, at least three phosphate acquisition genes, including a cell surface acid phosphatase Pho1, an inorganic phosphate transporter Pho84, and a glycerophosphate transporter Tgp1, have been identified to responsible for phosphate metabolisms (Carter-O’Connell et al., 2012). At least three lncRNAs have been reported to be transcribed under phosphate-rich conditions and repressed upon starvation, and they repress their sense-oriented target genes in cis through transcription interference (Ard et al., 2014; Chatterjee et al., 2016; Garg et al., 2018). The lncRNA Pho1-Repressing Transcript (prt) arises from the 1,147 nt upstream of the pho1 mRNA transcription start site in response to the addition of phosphate. However, with the deprivation of phosphate, the expression of prt is abolished through an unknown mechanism, leading to the expression of pho1 (Chatterjee et al., 2016). The lncRNA nc-tgp1, which originates at 1,823 nt upstream of the tgp1 mRNA start site, is transcribed in the presence of phosphate. However, upon phosphate deprivation, the derepression of nc-tgp1 leads to a decrease in the nucleosome occupancy, then result in the expression of tgp1 (Ard et al., 2014). The expression of both pho1 and tgp1 mRNAs depends on the DNA-binding transcription factor Pho7. It can recognize a 12 nt sequence motif (5′- TCG(G/C)(A/T)xxTTxAA-3′) present in the pho1 and tgp1 promoters (Schwer et al., 2017; Garg et al., 2019). The transcription of prt and nc-tgp1 may increase nucleosome density at the pho1 and tgp1 promoters, respectively, and displace Pho7 from the respective promoters (Figure 2A). Recently, it was reported that the dissociation of Pho7 from the pho1 locus which results in the transcription of prt was governed not only by the lncRNA prt itself but also by RNAPII, depending on its phosphorylation status (Chatterjee et al., 2016). Changes in the phosphorylation status of RNAPII may lead to the early termination of prt transcription, resulting in the derepression of pho1. A similar mechanism has been described for nc-tgp1 and its target gene tgp1 (Ard et al., 2014; Sanchez et al., 2018). However, in addition to prt, another regulatory RNA, named prt2, controls the expression of pho84 (Garg et al., 2018). Similar to the previously mentioned lncRNAs, prt2 is transcribed upon phosphate starvation and regulate the repression of its neighbor gene pho84 (Figure 2B). Moreover, the phosphorylation status of RNAPII impacts the expression of its target gene. Interestingly, prt2 not only affects its adjacent gene pho84, but also influence the expression of the lncRNA prt and its regulated gene pho1. The inactivation of prt2 leads to an upregulation of pho84, which consequently results in a downregulation of prt, and finally conduct to the transcription of pho1 (Garg et al., 2018). Both prt and nc-tgp1 are unstable owing to degradation by the nuclear exosome, and both carry a cluster of Mmi1-binding DSR (determinant of selective removal) motifs. The binding of Mmi1 in prt and nc-tgp1 induces the assembly of heterochromatin at their target gene loci (Shah et al., 2014). However, the specific contributions of Mmi1 and the nuclear exosome involved in transcription interference remain to be elucidated. However, this mechanism is independent of the regulatory impacts on their adjacent genes (Ard et al., 2014; Chatterjee et al., 2016).
As in S. pombe, a short antisense transcript of PHO84 also have been reported to affect the expression of PHO84 in S. cerevisiae. The antisense transcript, which originates from PHO84 locus, accumulates during aging, leading to the recruitment of the histone deacetylase complex Hda1/2/3 to the PHO84 promoter, the deacetylation of H3K18, and the silencing of PHO84 sense transcription (Camblong et al., 2007). These transcripts are degraded rapidly by the nuclear exosome subunit Rrp6. In ΔRrp6 cells during phosphate starvation, the induction of PHO84 is delayed because of the lncRNA accumulation (Castelnuovo et al., 2013).
Interestingly, although the majority of antisense transcripts are generally involved in the anti-regulation of their sense strands, there are still rare examples of positive gene regulation through antisense transcripts. For example, the expression of PHO5 is induced during phosphate starvation and shut-off in the presence of phosphate. A 2.4 kb antisense lncRNA activates PHO5 transcription during phosphate starvation (Uhler et al., 2007). This lncRNA is initiated at the 3′ end of the PHO5 gene, and it spans the PHO5 open reading frame and its promoter region. This antisense transcript is repressed during phosphate starvation. The low expression level of the ncRNA allows histone eviction from the PHO5 promoter and subsequent RNAPII recruitment, which enhances the initiation of PHO5 transcription (Uhler et al., 2007).
Galactose Utilization
The GAL1-10 cluster of S. cerevisiae is tightly regulated by environmental sugar availability. In the presence of galactose and the absence of glucose, GAL genes are induced, while in the presence of glucose, they are repressed. This highly regulated nutrient-response system allows S. cerevisiae to thrive on a wide range of carbon sources. At least two lncRNAs have been characterized as being involved in galactose metabolism by regulating the GAL cluster of genes. In 2008, Houseley et al. (2008) identified an antisense transcript GAL10-ncRNA that was initiated near the 3′ end of GAL10 and terminated in the GAL1-coding region in the absence of galactose (a condition that represses GAL10). The transcription of this antisense transcript is associated with the transcriptional activator Reb1, which binds to chromatin near the site of lncRNA initiation (Figure 2C). The transcription of GAL10-ncRNA also needs the recruitment of Set2 methyltransferase and histone deacetylation activities in cis, leading to increased H3K4 di- and tri-methylation within the GAL10-coding region, increased H3K36 tri-methylation, and decreased H3 acetylation across the GAL1-10 loci. This results in the silencing of all GAL loci (Houseley et al., 2008; Pinskaya et al., 2009). In addition, in the absence of galactose, although the Gal4 activator constitutively binds to GAL promoters, its activity is suppressed by Gal80 (Selleck and Majors, 1987). However, in the presence of galactose, GAL gene is activated with the release of Gal4. Both the GAL3 and GAL1 genes are activated by the Gal4 activator, forming two positive-feedback loops and leading to stable Gal4 release (Zenke et al., 1996; Acar et al., 2005; Venturelli et al., 2012). Furthermore, RNA degradation mechanisms also affect the expression of GAL10-ncRNA (Yoon et al., 2010; Figure 2C). In addition, the level of GAL10-lncRNA expression is elevated in the absence of the decapping enzyme Dcp2, and the degradation of GAL10-ncRNA is dependent on the cytoplasmic and nuclear 5’–3’ exonucleases Xrn1 and Rat1, respectively (Geisler et al., 2012). However, the deletion of Xrn1 has a limited impact on GAL1 expression. Another lncRNA termed GAL4-lncRNA has also been reported to regulate the expression of the transcriptional activator GAL4, although its mechanism has not been clarified in detail (Geisler et al., 2012).
Serine Utilization
In media containing serine, the lncRNA SRG1 is transcribed under serine-rich conditions, and it regulates the expression of its adjacent SER3 gene that encodes a phosphoglycerate dehydrogenase involved in serine biosynthesis (Martens et al., 2004). This is the first example of non-coding transcriptional interference in yeast. When serine is available to the cells, SRG1 transcription is turned on through a serine-dependent activator, Cha4, that recognizes an upstream activating sequence (UAS) in the SRG1 promoter (Martens et al., 2005). Moreover, a coactivator complex, namely SAGA, and the ATPase subunit of the Swi/Snf chromatin-remodeling complex is recruited to initiate SRG1 transcription, leading to the derepression of SER3 by increasing nucleosome occupancy at the SER3 promoter (Martens et al., 2005; Figure 2D). Recently, it was revealed that the transcription of SRG1 also requires the involvement of the transcription elongation complex FACT (Facilitates Chromatin Transcription), the Paf1 complex, chromatin reassembly factors (Spt6, Spn1, and Spt2), and specific amino acids in histones H3 and H4 (Martens et al., 2005; Pruneski et al., 2011; Thebault et al., 2011; Hainer et al., 2012; Hainer and Martens, 2016). In the absence of FACT and Spt6/Spn1, the density of nucleoso—mes over the SER3 promoter region was decreased but have no impact on SRG1 transcription (Hainer et al., 2011). In the absence of serine, although the Cha4 is constitutively bound to the lncRNA promoter, its transcription is repressed and the SER3 is depleted of nucleosomes, allowing both TBP and RNAPII and/or other unknown activator to bind and activate SER3 transcription (Figure 2D; Hainer et al., 2011).
Glucose Starvation
In the fission yeast S. pombe, glucose starvation induces the expression of the fbp1 gene that encodes a fructose-1,6-bis phosphatase (Hirota et al., 2008). There are two cis-acting regulatory elements in the upstream region of fbp1: UAS1 and UAS2. During glucose starvation, UAS1 recruits the transcription factor Atf1 and then induces the expression of non-coding transcripts upstream of the fbp1 gene, the so-called mlonRNAs (Figure 2E). These metabolic stress-induced lncRNAs allows the zinc-finger transcription factor Rst2 binds to UAS2 by disrupting the chromatin structure around UAS2 (Hirota et al., 2008; Figure 2E). The chromatin remodeling around the TATA box of the fbp1 promoter results in the full expression of fbp1 in the absence of glucose (Hirota et al., 2008). In addition, both the mlonRNAs and their antisense transcripts are degraded by the nuclear exosome/Rrp6 complex (Galipon et al., 2013; Miki et al., 2016).
Nitrogen Starvation
In S. cerevisiae, ASP3 encodes an enzyme Asparaginase II that can hydrolyze both D- and L-asparagine to aspartate and ammonium cations (Dunlop et al., 1978). ASP3 is activated in a nitrogen-limited environment (Dunlop et al., 1980; Kim et al., 1988). The lncRNA ncASP3, an intragenic sense-oriented transcript within the ASP3 coding region, is expressed when nitrogen is either available or depleted. The continuous expression of ncASP3 results in a high level of trimethylation of histone H3 at lysine 4 (H3K4me3) at the ASP3 promoter and makes this region more accessible for RNAPII’s transcription (Huang et al., 2010).
Cell-Cell Adhesion
In S. cerevisiae, the FLO11 gene encodes a cell wall glycoprotein that controls cell-cell adhesion, and only cells expressing FLO11 can undergo pseudohyphal growth (Halme et al., 2004). Research showed that the expression of the FLO11 gene is indirectly controlled by two lncRNAs, ICR1 and PWR1, forming a complicated regulatory model of cis-acting lncRNAs in fungi (Figure 2F). The lncRNA ICR1 (3.2 kb) is transcribed from the upstream intergenic region of FLO11 in the same direction while PWR1 (1.2 kb) is antisense to ICR1. The expression of FLO11 is repressed when the transcription factor Sfl1 binds to the FLO11 promoter, while another transcription factor, Flo8, activates it. Both Flo8 and Sfl1 compete for FLO11 promoter binding, and which of the two lncRNAs is expressed is determined through histone deacetylase Hda1 regulation. Hda1-mediated deacetylation condenses the chromatin at the Flo8- and Sfl1-binding sites. When Flo8 binds Sfl1, the transcription of PWR1 is activated. Then, PWR1 interferes with ICR1 through transcriptional interference, leading to the expression of FLO11. Conversely, the expression of ICR1 is activated with the binding of Sfl1 and the recruitment of histone deacetylase Hda1, then prevent the transcription of FLO11 (Figure 2F; Bumgarner et al., 2009).
Circadian Rhythm Maintenance
In N. crassa, the circadian clock core regulatory gene frq generates sustained rhythmicity (Gardner and Feldman, 1980). The long non-coding qrf, the antisense transcript of frq, is required for the rhythmic conidiation of the fungus. The transcription of qrf affects the clock’s response to light through chromatin modifications at the frq promoter (Kramer et al., 2003; Belden et al., 2011). The transcription of qrf represses frq expression by mediating chromatin modifications and the prior termination of transcription, and it regulates clock resetting. However, frq transcription also inhibits qrf expression and drives the antiphasic expression of qrf. Thus, the transcription of frq and qrf forms mutual inhibition of a double-negative feedback loop that is interconnected with the core feedback loop (Xue et al., 2014).
Stress Response
In S. cerevisiae, the modulation of cell-cycle control is controlled by the stress-activated protein kinase p38/Hog1. Upon osomostress, a large set of lncRNAs, including the antisense lncRNA of CDC28, is induced by Hog1 to regulate the expression of the master cell cycle regulator CDK1/Cdc28 (Nadal-Ribelles et al., 2014). Through increasing the Cdc28 level, the CDC28 lncRNA promotes the stressed cells efficient enter into the cell cycle. The antisense lncRNA is induced when Hog1 is associated with the 3′ region of CDC28. Then forms a gene looping between the 5′- and 3′- UTRs of CDC28, which causes the relocation of Hog1 to the 5′ region. The relocated Hog1 then promotes chromatin remodeling by recruiting the RNA-induced silencing remodeling complex and inducing CDC28 expression (Nadal-Ribelles et al., 2014).
In addition, another lncRNASPNCRNA.1164 has also been reported to regulate the expression of a stress-responsive transcription factor, Atf1, under oxidative stress conditions, although its mode of action remains unknown (Leong et al., 2014). Glucose starvation cannot induce the expression of SPNCRNA.1164, suggesting that it may be respond only to specific stress stimuli (Leong et al., 2014).
Telomere Maintenance
In S. cerevisiae, two sorts of telomere-associated lncRNAs have been identified: TERRA and the telomerase RNA TCL1. TERRA is an evolutionary conserved lncRNA that has been found in many eukaryotic cells, such as those of humans (Homo sapiens), budding yeast (S. cerevisiae), fission yeast (S. pombe), mice (Mus musculus), zebrafish (Danio rerio), and plants (Arabidopsis thaliana), indicating its important functions in regulating telomerase (Azzalin et al., 2007; Luke et al., 2008; Azzalin and Lingner, 2015). During the telomeric synthesis process, TERRA forms a scaffold that connects both the telomeric DNAs and chromatin-modifying enzymes to maintain telomerase activity (Luke et al., 2008). In addition, TERRA interacts with other associated telomeric proteins to regulate the integrity of the telomere. In yeast, TERRA is regulated by the 5′–3′ exonuclease, Rat1p and stabilized by Pap1p. In the mutant rat1-1 cells, TERRA is accumulated and harbored in short telomeres because of defects in telomerase-mediated telomere elongation (Cusanelli and Chartrand, 2015).
Another telomere-associated lncRNA described in S. cerevisiae is TLC1. This lncRNA is transcribed on chromosome II and has a secondary structure composed of three long arms (Singer and Gottschling, 1994). TLC1 form the telomerase complex by physically interacting with the proteins Est1, Est2, and Est3. This complex involves in the telomeric DNA repeats synthesis and prevent telomere-shortening cell division (Gallardo et al., 2008). In S. pombe, a homolog of TLC1, named TER1, has also been discovered. In comparison with TLC1, TER1 is much larger and contains more invariant repeats (Leonardi et al., 2008).
Pathogenicity
Although there only a few fungal lncRNAs related to pathogenicity has been reported, we still believe that lncRNAs derived from pathogenic fungi may play important roles during fungi-host interaction. These lncRNAs which is involved in the pathogenicity may enhance the virulence of fungi or they can repress host immune response during infection. In the biotrophic basidiomycete fungus Ustilago maydis, RT-PCR showed that ncrna1, an antisense transcript complementary to the 3′-UTR of um02150, is expressed during corn infection. The infected ability was reduced with the absence of ncrna1, suggesting that ncrna1 may be involved in pathogenesis though the mechanism has yet to be determined (Morrison and Saville, 2012). Also in U. maydis, the expression of the natural antisense transcript as-um02151 results in a twofold increase in complementary mRNA levels. The alteration of its expression decreases pathogenesis too (Donaldson and Saville, 2013). However, both detailed regulatory mechanisms of these two putative lncRNAs which may relate to pathogenesis have not been identified yet. Recently, Tang et al. elucidated the lncRNA profiles during infection and development of phytopathogen Ustilaginoidea virens. RNA-seq analyses revealed more than 1700 lncRNAs in U. virens (Tang et al., 2021). However, functions of these lncRNAs still undetermined yet.
Other Functions
In 2008, Berretta et al. found that several antisense ncRNAs were transcribed within Ty1 elements in S. cerevisiae, and function in trans (Berretta et al., 2008). The expression of those antisense ncRNA may conduct to the suppression of Ty1 elements in trans and also lead to the repression of the Ty1 promoter and 5’ long terminal repeat, which suggests that antisense ncRNAs regulate Ty1 in trans. These ncRNAs hold at very low levels because of the RNA surveillance mechanisms. Additionally, Set1 is required for the silencing of Ty1 elements by antisense Ty1 transcripts (Berretta et al., 2008). In S. cerevisiae, the protein-coding sense gene MDF1 significantly suppresses the mating efficiency in a rich medium by binding MATα2, thereby promoting vegetative growth. The antisense gene ADF1 acts as a transcriptional suppressor that regulates the expression of the sense gene MDF1 (Li et al., 2010).
In addition, besides the regulatory lncRNA which was functionally identified in yeast, several lncRNAs have also been characterized in filamentous fungi. For instance, the deletion of the poorly transcribed lncRNA carP (Fo-carP and Ff-carP in the fungi Fusarium oxysporum and F. fujikuroi, respectively) leads to strong reductions of the carotenoid biosynthesis genes at mRNA levels. However, the content of carS mRNA is maintained at a higher level, indicating that carP is involved in activating the expression of carotenoid genes in Fusarium with an unknown molecular mechanism (Parra-Rivero et al., 2020). Additionally, in the fungal pathogen Cryptococcus neoformans, the lncRNA RZE1 controls the yeast-to-hyphal transition by regulating the key morphogenesis regulator Znf2 (Chacko et al., 2015). Moreover, a lncRNA termed HAX1 has been identified as promoting cellulase expression in Trichoderma reesei (Till et al., 2018). In the maize pathogen Cochliobolus heterostrophus, the transcription factor Cmr1, which regulates melanin biosynthesis, is transcribed in both sense and antisense directions. The transition of the melanin gene cluster is regulated by both the antisense transcript of Cmr1 and the MAPK pathway, resulting in the euchromatin and heterochromation (Eliahu et al., 2007).
Conclusion and Perspectives
With the availability of high-throughput sequencing technology, the identification of lncRNAs is becoming easier. Thus, scientists are facing another major challenge: the functional characterization of the regulatory mechanisms of ncRNAs. However, owing to the non-conservative, development- and condition-specific nature of lncRNAs, it is still difficult to identify their functions at the molecular level. For example, although the sporulation processes of the two yeasts S. cerevisiae and S. pombe are both controlled by lncRNA-based chromatin silencing, their regulatory mechanisms are totally divergent. This suggested that there is great diversity in regulatory mechanisms in fungi kingdom. Therefore, more extensive studies are still needed to clarify the molecular mechanisms of expression, regulation, and functions of lncRNAs. Moreover, functional identifications in other fungi, especially pathogenic fungi, will result in a comprehensive understanding of pathogenicity mechanisms in fungi. However, pathogenicity-related lncRNAs in fungi is still less well documented. Advanced sequencing data will unveil profiles of lncRNAs and provide new insights and promising lncRNA candidates in this area.
Recently, a new type of lncRNA, circRNA, has been identified as playing important roles in animals and plants (Kristensen et al., 2019), but little attention has been focused on fungal circRNAs. Although Yuan et al. (2018) identified more than 8,000 circRNAs from the model plant-pathogenic fungus M. oryzae, and these circRNAs may play important roles in the regulation of growth and development. Thus, there is enormous potential for increasing our understanding of the relatively new field of circRNAs in fungi.
We believe that ongoing fungal lncRNA-based studies will provide important insights into how lncRNAs regulate important biological processes in fungi. Research in simple eukaryotes may provide important clues for understanding how this novel discovered regulatory lncRNAs function in eukaryotic cells.
Author Contributions
JL conceived the manuscript. JL, XL, ZY, ZH, and K-QZ discussed and developed its conceptual framework. JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for their valuable suggestions. We thank Lesley Benyon, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Footnotes
Funding. This work was jointly funded by the National Natural Science Foundation of China (approved nos. 31970073, 31760538, and 31560025).
References
- Acar M., Becskei A., van Oudenaarden A. (2005). Enhancement of cellular memory by reducing stochastic transitions. Nature 435 228–232. 10.1038/nature03524 [DOI] [PubMed] [Google Scholar]
- Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160 595–606. 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Watanabe H., Lipman D. J., Koonin E. V. (2000). Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 97 11319–11324. 10.1073/pnas.200346997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R., Tong P., Allshire R. C. (2014). Long non-coding RNA-mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 5:5576. 10.1038/ncomms6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55 383–396. 10.1016/j.molcel.2014.06.011 [DOI] [PubMed] [Google Scholar]
- Ariel F., Lucero L., Christ A., Mammarella M. F., Jegu T., Veluchamy A., et al. (2020). R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell 77 1055–1065.e4. 10.1016/j.molcel.2019.12.015 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Lingner J. (2015). Telomere functions grounding on TERRA firma. Trends Cell Biol. 25 29–36. 10.1016/j.tcb.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Belden W. J., Lewis Z. A., Selker E. U., Loros J. J., Dunlap J. C. (2011). CHD1 Remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet. 7:e1002166. 10.1371/journal.pgen.1002166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J., Pinskaya M., Morillon A. (2008). A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 22 615–626. 10.1101/gad.458008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Ballabio A., Rupert J. L., Lafreniere R. G., Grompe M., Tonlorenzi R., et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349 38–44. 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R. (2009). Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 106 18321–18326. 10.1073/pnas.0909641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S. M. (2007). MicroRNA functions. Annu. Rev. Cell Dev. Bi. 23 175–205. 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L. M., Stutz F. (2009). Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 23 1534–1545. 10.1101/gad.522509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F. (2007). Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131 706–717. 10.1016/j.cell.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Cao M., Zhao J., Hu G. (2019). Genome-wide methods for investigating long noncoding RNAs. Biomed. Pharmacother 111 395–401. 10.1016/j.biopha.2018.12.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-O’Connell I., Peel M. T., Wykoff D. D., O’Shea E. K. (2012). Genome-Wide characterization of the phosphate starvation response in schizosaccharomyces pombe. BMC Genomics 13:697. 10.1186/1471-2164-13-697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Sontheimer E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136 642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnuovo M., Rahman S., Guffanti E., Infantino V., Stutz F., Zenklusen D. (2013). Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat. Struct. Mol. Biol. 20 851–858. 10.1038/nsmb.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko N., Zhao Y., Yang E., Wang L., Cai J. J., Lin X. (2015). The lncRNA RZE1 controls cryptococcal morphological transition. PLoS Genet 11:e1005692. 10.1371/journal.pgen.1005692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Sanchez A. M., Goldgur Y., Shuman S., Schwer B. (2016). Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA 22 1011–1025. 10.1261/rna.056515.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S. W., Gruhl F., Mattick J. S., Dinger M. E. (2013). Long noncoding RNAs and the genetics of cancer. Br. J. Cancer 108 2419–2425. 10.1038/bjc.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie K. R., Weng S., Balakrishnan R., Costanzo M. C., Dolinski K., Dwight S. S., et al. (2004). Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32 D311–D314. 10.1093/nar/gkh033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M. D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E., et al. (1994). enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanelli E., Chartrand P. (2015). Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 6:143. 10.3389/fgene.2015.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Hannon G. J. (2011). Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12 19–31. 10.1038/nrg2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C. J., Bofkin L., et al. (2006). A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 103 5320–5325. 10.1073/pnas.0601091103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. A., Ares M. (2006). Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 103 3262–3267. 10.1073/pnas.0507783103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D. Q., Okamasa K., Yamane M., Tsutsumi C., Haraguchi T., Yamamoto M., et al. (2012). Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in Meiosis. Science 336 732–736. 10.1126/science.1219518 [DOI] [PubMed] [Google Scholar]
- Donaldson M. E., Saville B. J. (2012). Natural antisense transcripts in fungi. Mol. Microbiol. 85 405–417. 10.1111/j.1365-2958.2012.08125.x [DOI] [PubMed] [Google Scholar]
- Donaldson M. E., Saville B. J. (2013). Ustilago maydis natural antisense transcript expression alters mRNA stability and pathogenesis. Mol. Microbiol. 89 29–51. 10.1111/mmi.12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., Fink G. R., Bartel D. P. (2011). Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333:1592. 10.1126/science.1209575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., Weinberg D. E., Xie K. T., Mower J. P., Wolfe K. H., Fink G. R., et al. (2009). RNAi in budding yeast. Science 326 544–550. 10.1126/science.1176945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop P. C., Meyer G., Ban D., Roon R. J. (1978). Characterization of two forms of asparaginase in Saccharomyces cerevisiae. J. Biol. Chem. 253 1297–1304. [PubMed] [Google Scholar]
- Dunlop P. C., Meyer G. M., Roon R. J. (1980). Nitrogen catabolite repression of asparaginase II in Saccharomyces cerevisiae. J. Bacteriol. 143 422–426. 10.1128/jb.143.1.422-426.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliahu N., Igbaria A., Rose M. S., Horwitz B. A., Lev S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot Cell 6 421–429. 10.1128/ec.00264-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Bozzoni I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15 7–21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Fraga de Andrade I., Mehta C., Bresnick E. H. (2020). Post-transcriptional control of cellular differentiation by the RNA exosome complex. Nucleic Acids Res. 48 11913–11928. 10.1093/nar/gkaa883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Galipon J., Miki A., Oda A., Inada T., Ohta K. (2013). Stress-induced lncRNAs evade nuclear degradation and enter the translational machinery. Genes Cells 18 353–368. 10.1111/gtc.12042 [DOI] [PubMed] [Google Scholar]
- Gallardo F., Olivier C., Dandjinou A. T., Wellinger R. J., Chartrand P. (2008). TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 27 748–757. 10.1038/emboj.2008.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner G. F., Feldman J. F. (1980). The frq locus in Neurospora crassa: a key element in circadian clock organization. Genetics 96 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Goldgur Y., Sanchez A. M., Schwer B., Shuman S. (2019). Structure of fission yeast transcription factor Pho7 bound to pho1 promoter DNA and effect of Pho7 mutations on DNA binding and phosphate homeostasis. Mol. Cell Biol. 39 e00132–19. 10.1128/mcb.00132-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Sanchez A. M., Shuman S., Schwer B. (2018). A long noncoding (lnc)RNA governs expression of the phosphate transporter Pho84 in fission yeast and has cascading effects on the flanking prt lncRNA and pho1 genes. J. Biol. Chem. 293 4456–4467. 10.1074/jbc.RA117.001352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8 113–126. 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- Geisler S., Lojek L., Khali A. M., Baker K. E., Coller J. (2012). Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell 45 279–291. 10.1016/j.molcel.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand B., Mead J., Bruning A., Apostolopoulos N., Tadigotla V., Nagaraj V., et al. (2011). Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol. Cell. Biol. 31 1701–1709. 10.1128/mcb.01071-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10 94–108. 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Kugel J. F. (2006). Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 7 612–616. [DOI] [PubMed] [Google Scholar]
- Gudipati R. K., Xu Z., Lebreton A., Séraphin B., Steinmetz L. M., Jacquier A., et al. (2012). Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell 48:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Esteller M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19:1068. [DOI] [PubMed] [Google Scholar]
- Guttman M., Rinn J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482 339–346. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Charsar B. A., Cohen S. B., Martens J. A. (2012). Identification of mutant versions of the Spt16 histone chaperone that are defective for transcription-coupled nucleosome occupancy in Saccharomyces cerevisiae. G3 (Bethesda) 2 555–567. 10.1534/g3.112.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Martens J. A. (2016). Regulation of chaperone binding and nucleosome dynamics by key residues within the globular domain of histone H3. Epigenetics Chrom. 9:17. 10.1186/s13072-016-0066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A. (2011). Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25 29–40. 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R. (2004). Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116 405–415. 10.1016/s0092-8674(04)00118-7 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Hirota K., Miyoshi T., Kugou K., Hoffman C. S., Shibata T., Ohta K. (2008). Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456 130–134. 10.1038/nature07348 [DOI] [PubMed] [Google Scholar]
- Hongay C. F., Grisafi P. L., Galitski T., Fink G. R. (2006). Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127 735–745. 10.1016/j.cell.2006.09.038 [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. (2006). RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7 529–539. 10.1038/nrm1964 [DOI] [PubMed] [Google Scholar]
- Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. (2008). A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32 685–695. 10.1016/j.molcel.2008.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. M., Liu T. Y., Shang C. L., Zhao Y. H., Wang W., Liang Y. C., et al. (2018). Identification of lncRNAs by microarray analysis reveals the potential role of lncRNAs in cervical cancer pathogenesis. Oncol. Lett. 15 5584–5592. 10.3892/ol.2018.8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-C., Chen H.-T., Teng S.-C. J. R. (2010). Intragenic transcription of a noncoding RNA modulates expression of ASP3 in budding yeast. RNA 16 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Chang H. Y. (2010). Long noncoding RNA in genome regulation Prospects and mechanisms. RNA Biol. 7 582–585. 10.4161/rna.7.5.13216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43 621–629. 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A. (2009). The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 10 833–844. 10.1038/nrg2683 [DOI] [PubMed] [Google Scholar]
- Januszyk K., Lima C. D. (2010). “Structural components and architectures of RNA exosomes,” in Rna Exosome, ed. Jensen T. H. (New York, NY: Springer New York; ), 9–28. [Google Scholar]
- Kim D. H., Sung S. (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40 302–312.e4. 10.1016/j.devcel.2016.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. W., Kamerud J. Q., Livingston D. M., Roon R. J. (1988). Asparaginase II of saccharomyces cerevisiae. characterization of the ASP3 gene. J. Biol. Chem. 263 11948–11953. [PubMed] [Google Scholar]
- Kornienko A. E., Guenzl P. M., Barlow D. P., Pauler F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. 10.1186/1741-7007-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C., Loros J. J., Dunlap J. C., Crosthwaite S. K. (2003). Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421 948–952. 10.1038/nature01427 [DOI] [PubMed] [Google Scholar]
- Kristensen L. S., Andersen M. S., Stagsted L. V. W., Ebbesen K. K., Hansen T. B., Kjems J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 675–691. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- Kung J. T. Y., Colognori D., Lee J. T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193 651–669. 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenois A., Liu Y. C., Walther T., Chalmel F., Evrard B., Granovskaia M., et al. (2011). Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. U.S.A. 108 1058–1063. 10.1073/pnas.1016459108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T. (2012). Epigenetic regulation by long noncoding RNAs. Science 338 1435–1439. 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- Leonardi J., Box J. A., Bunch J. T., Baumann P. (2008). TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 15 26–33. 10.1038/nsmb1343 [DOI] [PubMed] [Google Scholar]
- Leong H. S., Dawson K., Wirth C., Li Y. Y., Connolly Y., Smith D. L., et al. (2014). A global non-coding RNA system modulates fission yeast protein levels in response to stress. Nat. Commun. 5:3947. 10.1038/ncomms4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Dong Y., Jiang Y., Jiang H., Cai J., Wang W. (2010). A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 20 408–420. 10.1038/cr.2010.31 [DOI] [PubMed] [Google Scholar]
- Luke B., Panza A., Redon S., Iglesias N., Li Z. J., Lingner J. (2008). The Rat1p 5 ’ to 3 ’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32 465–477. 10.1016/j.molcel.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Brodersen D. E., Jensen T. H. (2009). Origins and activities of the eukaryotic exosome. J. Cell Sci. 122 1487–1494. 10.1242/jcs.047399 [DOI] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F. (2004). Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429 571–574. 10.1038/nature02538 [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F. (2005). Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19 2695–2704. 10.1101/gad.1367605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Guttman M. (2019). Approaches for understanding the mechanisms of long noncoding RNA regulation of gene expression. Cold Spring Harb. Perspect. Biol. 11:a032151. 10.1101/cshperspect.a032151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki A., Galipon J., Sawai S., Inada T., Ohta K. (2016). RNA decay systems enhance reciprocal switching of sense and antisense transcripts in response to glucose starvation. Genes Cells 21 1276–1289. 10.1111/gtc.12443 [DOI] [PubMed] [Google Scholar]
- Mishra K., Kanduri C. (2019). Understanding long noncoding RNA and chromatin interactions: what we know so far. Non Coding RNA 5:54. 10.3390/ncrna5040054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Bühler M., Buker S. M., Colmenares S. U., Gerace E. L., Gerber S. A., et al. (2006). Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harb. Symp. Quant. Biol. 71 461–471. 10.1101/sqb.2006.71.044 [DOI] [PubMed] [Google Scholar]
- Morrison E. N., Saville B. J. (2012). Identification and analysis of genes expressed in the Ustilago maydis dikaryon: uncovering a novel class of pathogenesis genes. Can. J. Plant Pathol. 34 417–435. [Google Scholar]
- Murata T., Kadotani N., Yamaguchi M., Tosa Y., Mayama S., Nakayashiki H. (2007). siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae. Nucleic Acids Res. 35 5987–5994. 10.1093/nar/gkm646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal-Ribelles M., Sole C., Xu Z., Steinmetz L. M., de Nadal E., Posas F. (2014). Control of Cdc28 CDK1 by a stress-induced lncRNA. Mol. Cell 53 549–561. 10.1016/j.molcel.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., et al. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320 1344–1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T., Fraser P. (2011). No-nonsense functions for long noncoding RNAs. Cell 145 178–181. 10.1016/j.cell.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Nagarajan V. K., Jones C. I., Newbury S. F., Green P. J. (2013). XRN 5 ’ -> 3 ’ exoribonucleases: structure, mechanisms and functions. Biochimica Biophys. Acta Gene Regul. Mech. 1829 590–603. 10.1016/j.bbagrm.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair L., Chung H., Basu U. (2020). Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat. Rev. Mol. Cell Biol. 21 123–136. 10.1038/s41580-019-0209-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H., Malabat C., D’Aubenton-Carafa Y., Xu Z., Steinmetz L. M., Jacquier A. (2009). Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou M. D., Hatzigeorgiou A. G. (2016). Analyzing MiRNA-LncRNA interactions. Methods Mol. Biol. 1402 271–286. 10.1007/978-1-4939-3378-5_21 [DOI] [PubMed] [Google Scholar]
- Parra-Rivero O., Pardo-Medina J., Gutiérrez G., Limón M. C., Avalos J. (2020). A novel lncRNA as a positive regulator of carotenoid biosynthesis in Fusarium. Sci. Rep. 10:678. 10.1038/s41598-020-57529-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E. (2012). NON-CODING RNA MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13 271–282. 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- Pefanis E., Wang J. G., Rothschild G., Lim J., Kazadi D., Sun J. B., et al. (2015). RNA Exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161 774–789. 10.1016/j.cell.2015.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M., Gourvennec S., Morillon A. (2009). H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28 1697–1707. 10.1038/emboj.2009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskol R., Ramaswami G., Li J. B. (2013). Reliable identification of genomic variants from RNA-Seq Data. Am. J. Hum. Genet. 93 641–651. 10.1016/j.ajhg.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneski J. A., Hainer S. J., Petrov K. O., Martens J. A. (2011). The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot Cell 10 1283–1294. 10.1128/ec.05141-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiah M., Shum, Eleen Y., Wilkinson, Miles F. (2012). How to activate a gene: decap its associated noncoding RNA. Mol. Cell 45 271–273. 10.1016/j.molcel.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81 145–166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M. P., Tongprasit W., Sethi H., Chin C. S., Stolc V. (2006). Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc. Natl. Acad. Sci. U.S.A. 103 4192–4197. 10.1073/pnas.0507669103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. M., Shuman S., Schwer B. (2018). Poly(A) site choice and Pol2 CTD Serine-5 status govern lncRNA control of phosphate-responsive tgp1 gene expression in fission yeast. RNA 24 237–250. 10.1261/rna.063966.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mejias A., Tay Y. (2015). Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J. Hematol. Oncol. 8:30. 10.1186/s13045-015-0129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jensen T. H. (2008). The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 33 501–510. 10.1016/j.tibs.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Schulz D., Schwalb B., Kiesel A., Baejen C., Torkler P., Gagneur J., et al. (2013). Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155 1075–1087. 10.1016/j.cell.2013.10.024 [DOI] [PubMed] [Google Scholar]
- Schwer B., Sanchez A. M., Garg A., Chatterjee D., Shuman S. (2017). Defining the DNA binding site recognized by the fission yeast Zn(2)Cys(6) transcription factor Pho7 and its role in phosphate homeostasis. mBio 8 e01218–17. 10.1128/mBio.01218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. E. (1987). In vivo DNA-binding properties of a yeast transcription activator protein. Mol. Cell Biol. 7 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Wittmann S., Kilchert C., Vasiljeva L. (2014). lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 28 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y., Otsubo Y., Yamamoto M., Yamashita A. (2020). Meiotic gene silencing complex MTREC/NURS recruits the nuclear exosome to YTH-RNA-binding protein Mmi1. PLoS Genet. 16:e1008598. 10.1371/journal.pgen.1008598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y., Yamashita A., Yamamoto M. (2014). Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 4:140022. 10.1098/rsob.140022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. (2020). Transcriptional interference at tandem lncRNA and protein-coding genes: an emerging theme in regulation of cellular nutrient homeostasis. Nucleic Acids Res. 48 8243–8254. 10.1093/nar/gkaa630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E. (1994). TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266 404–409. 10.1126/science.7545955 [DOI] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 799–802. 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Tang J., Chen X., Yan Y., Huang J., Luo C., Tom H., et al. (2021). Comprehensive transcriptome profiling reveals abundant long non-coding RNAs associated with development of the rice false smut fungus, Ustilaginoidea virens. Environ. Microbiol. 10.1111/1462-2920.15432 [Epup ahead of print], [DOI] [PubMed] [Google Scholar]
- Thebault P., Boutin G., Bhat W., Rufiange A., Martens J., Nourani A. (2011). Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell Biol. 31 1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D. W., Dinger M. E. (2016). Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17 272–283. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- Till P., Pucher M. E., Mach R. L., Mach-Aigner A. R. (2018). A long noncoding RNA promotes cellulase expression in Trichoderma reesei. Biotechnol. Biofuels 11:78. 10.1186/s13068-018-1081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toesca I., Nery C. R., Fernandez C. F., Sayani S., Chanfreau G. F. (2011). Cryptic Transcription mediates repression of subtelomeric metal homeostasis genes. PLoS Genet. 7:e1002163. 10.1371/journal.pgen.1002163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler J. P., Herteit C., Svejstrup J. Q. (2007). A role for noncoding transcription in activation of the yeast PH05 gene. Proc. Natl. Acad. Sci. U.S.A. 104 8011–8016. 10.1073/pnas.0702431104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Chen C. L., d’Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., et al. (2011). XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475 114–117. 10.1038/nature10118 [DOI] [PubMed] [Google Scholar]
- van Werven F. J., Neuert G., Hendrick N., Lardenois A., Buratowski S., van Oudenaarden A., et al. (2012). Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150 1170–1181. 10.1016/j.cell.2012.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K. W., Ponting C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Li H., Gogol M. M., Workman J. L. (2016). Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat. Commun. 7:13610. 10.1038/ncomms13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli O. S., El-Samad H., Murray R. M. (2012). Synergistic dual positive feedback loops established by molecular sequestration generate robust bimodal response. Proc. Natl. Acad. Sci. U.S.A. 109 E3324–E3333. 10.1073/pnas.1211902109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth E. V., Januszyk K., Lima C. D. (2014). Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511 435–439. 10.1038/nature13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth E. V., Lima C. D. (2012). Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell 48 133–144. 10.1016/j.molcel.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., Sunwoo H., Spector D. L. (2009). Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23 1494–1504. 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Fang X., Zhu D., Dean C. (2020). Autonomous Pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol. 182 27–37. 10.1104/pp.19.01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., et al. (2005). Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121 725–737. 10.1016/j.cell.2005.04.030 [DOI] [PubMed] [Google Scholar]
- Xu Z. Y., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., et al. (2009). Bidirectional promoters generate pervasive transcription in yeast. Nature 457 1033–U1037. 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z. H., Ye Q. H., Anson S. R., Yang J. C., Xiao G., Kowbel D., et al. (2014). Transcriptional interference by antisense RNA is required for circadian clock function. Nature 514 650–653. 10.1038/nature13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeasmin F., Yada T., Akimitsu N. (2018). Micropeptides encoded in transcripts previously identified as long noncoding RNAs: a new chapter in transcriptomics and proteomics. Front. Genet. 9:144. 10.3389/fgene.2018.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Choi E. J., Parker R. (2010). Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189 813–827. 10.1083/jcb.200912019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. X., Yarrington R. M., Chuong E. B., Elde N. C., Stillman D. J. (2016). Disruption of promoter memory by synthesis of a long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 113 9575–9580. 10.1073/pnas.1601793113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. L., Wang Z., Xing J. J., Yang Q. Y., Chen X. L. (2018). Genome-wide Identification and characterization of circular RNAs in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 8:6757. 10.1038/s41598-018-25242-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F. T., Engles R., Vollenbroich V., Meyer J., Hollenberg C. P., Breunig K. D. (1996). Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272 1662–1665. 10.1126/science.272.5268.1662 [DOI] [PubMed] [Google Scholar]
- Zhang K., Fischer T., Porter R. L., Dhakshnamoorthy J., Zofall M., Zhou M., et al. (2011). Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331 1624–1627. 10.1126/science.1198712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Sun W., Guo Z., Zhang J., Yu H., Liu B. (2020). Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 254:116900. 10.1016/j.lfs.2019.116900 [DOI] [PubMed] [Google Scholar]
- Zhong Y. X., Du Y. J., Yang X., Mo Y. Z., Fan C. M., Xiong F., et al. (2018). Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 17:79. 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]