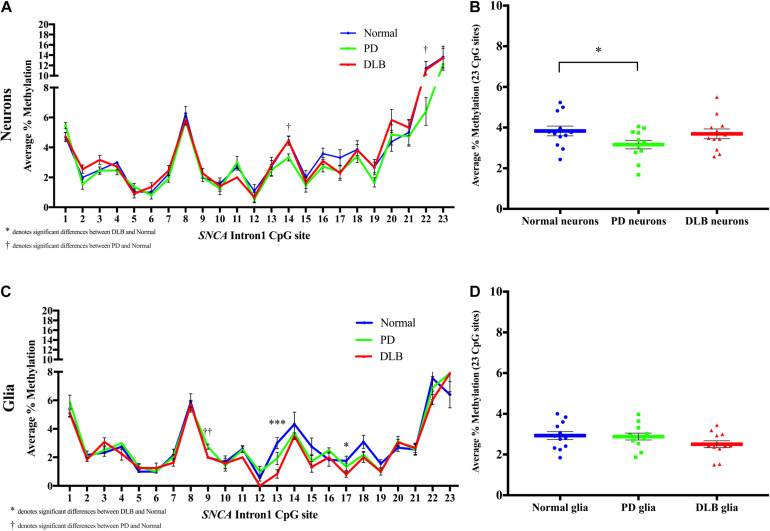
FIGURE 5.
DNA methylation profiles of the SNCA intron 1 CGI in the sorted neuronal and glia nuclei from frontal cortex tissues. DNA from each neuronal and glia sorted nuclei sample was bisulfite converted, and the methylation (%) of the individual CpGs was quantitatively determined by pyrosequencing. Graph depicts the average percentage of DNA methylation level at each of the 23 CpG sites within the SNCA intron 1 CGI for (A) neuronal nuclei and (C) glia nuclei from 20 PD, 20 DLB, and 20 matched normal control subjects. The significance of the differences in % of methylation was determined using one-way ANOVA followed by Dunnett’s post hoc test and denoted by asterisks (∗) for DLB vs normal, and by obelisks (†) for PD vs normal comparisons, with ∗/†p < 0.05, ∗∗/††p < 0.005, and ∗∗∗/†††p < 0.001. Box-and-scatterplot charts display the DNA methylation average percentages across all 23 CpG sites within the SNCA intron 1 CGI [Chr4: 89,836,150-89,836,593 (GRCh38/hg38)] for each (B) neuronal and (D) glia nuclei isolated and sorted from each subject (denoted by colored points). The box plots also depict the mean (denoted by colored horizontal lines) and upper and lower quartiles (denoted by gray thin lines) of DNA methylation average percentages across all 23 CpG sites for all (B) neuronal and (D) glia sorted nuclei in each group of PD, DLB, and normal control. The significance of the differences in % of methylation was determined using Student’s t-test and repeated using one-way ANOVA followed by Dunnett’s post hoc test. ∗indicates p < 0.05 for t-test; upon applying the Dunnett’s multiple comparisons method, the result did not reach statistical significance (p = 0.08).