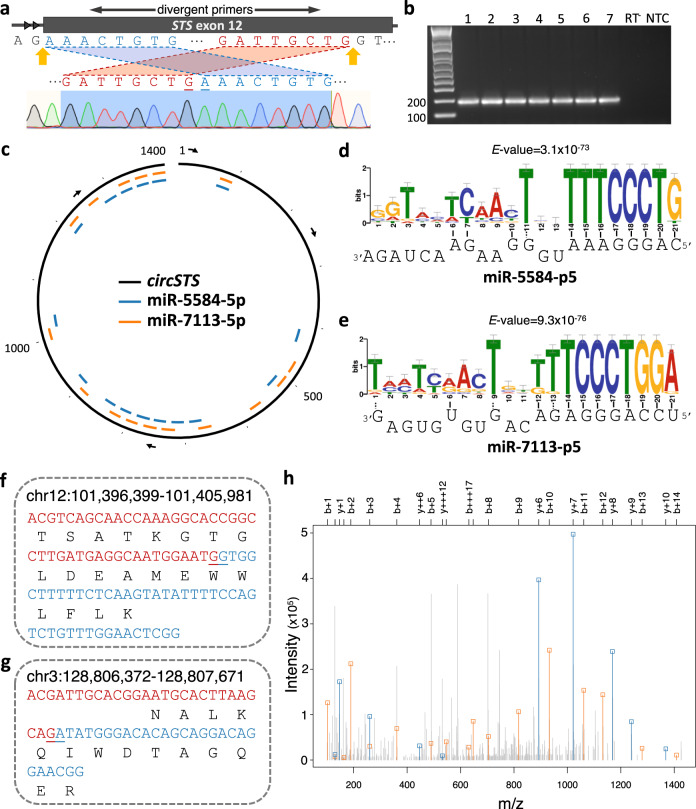
Fig. 3. circSTS as a putative miRNA sponge and evidence of peptides translated from circRNAs.
a The back-spliced positions (indicated in yellow arrows) are within the 12th exon (thick rectangle) of a lincRNA (ENST00000658154) encoded by STS. Back-splicing was assayed with divergent primers (top) and confirmed by Sanger sequencing (bottom). The first and last 8-bases, colored in blue (5’) and red (3’) respectively, are flanked by AG/GT (intronic acceptor/donor sites). b The expected size of the back-spliced PCR product from circSTS (170 bp) was validated by qPCR from seven placental samples. RT-: no reverse transcriptase; NTC: no template control. Source data are provided as a Source Data file. c circSTS (chrX:7,514,882-7,516,290) is represented as a black open circle with its putative binding sites for miR-5584-5p (blue) and miR-7113-5p (orange). Its relative base positions are marked for every 100th-base position and 1st, 500th, 1000th, and 1400th positions are numbered. The arrows indicate 5′-to-3′ direction. Sequence motif analyses of miR-5584-5p and miR-7113-p5 are shown in (d) and (e) respectively, with the corresponding sequence miRNAs shown at the bottom. The size of the sequence logo represents how well the binding base is conserved across the 16 predicted binding regions represented in (c). f–g Peptidomic evidence for translation of two circRNAs. The putative peptides are in black and the bases on either side of the BSJ are colored in red (5′) and blue (3′). The peptides were translated across the BSJ (indicated by underlines) of circRNAs. h Mass spectrum corresponding to the peptide sequence of (f). On the x-axis, the mass/charge ratio of the peptide fragments is shown, with the intensity on the y-axis. Peaks that can be attributed to b and y-ions (i.e., including the N- and C- termini respectively) of the peptide are indicated with a square and colored in red and blue respectively.