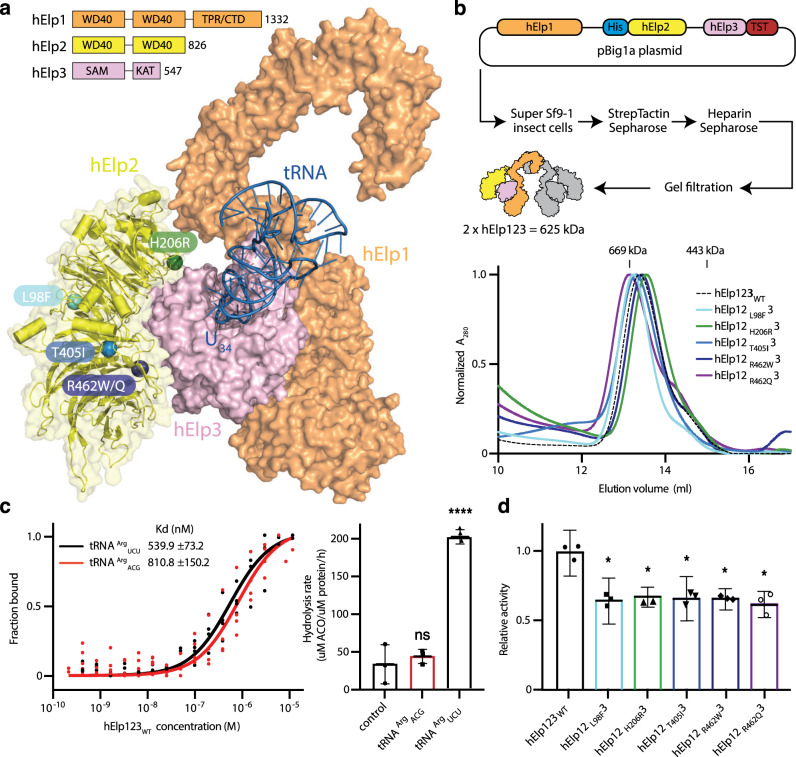
Fig. 3. Patient ELP2 mutations decrease Elongator activity.
a Human ELP123 (hElp123) domain organization scheme: TPR tetratricopeptide repeat domain, CTD C-terminal domain, SAM S-adenosyl methionine domain, KAT lysine acetyltransferase domain (top). Human ELP123 model with bound tRNA and depicted ELP2 mutations (bottom). Color code for molecules: hElp1—orange, hElp2—yellow, hElp3— pink, tRNA—blue. b Workflow scheme for hElp123 wild-type (hElp123WT) protein production and purification from insect cell expression system (top). Gel filtration profiles from Superose 6 Increase 10/300 GL column for all hElp123 variants (bottom; n = 3 independent experiments). c Microscale thermophoresis analysis of hElp123WT binding to tRNAArgUCU or tRNAArgACG with estimated dissociation constant values (Kd) and Kd fitting errors (left; mean ± SD; n = 4 independent measurements). Acetyl-CoA hydrolysis activity assay results for hElp123WT incubated with no tRNA (control), tRNAArgACG, or tRNAArgUCU (right; n = 3 independent experiments). d Normalized acetyl-CoA hydrolysis activity assay results for hElp123 variants incubated with tRNAArgUCU (n = 3 independent experiments). Statistical analysis: one-way ANOVA (α = 0.05) with a Dunnett’s post-hoc test. Statistically significant differences are indicated (*p ≤ 0.05; ****p ≤ 0.0001; ns not significant: p = 0.6823), Data represent mean ± SD. Source data are provided as a Source Data file.