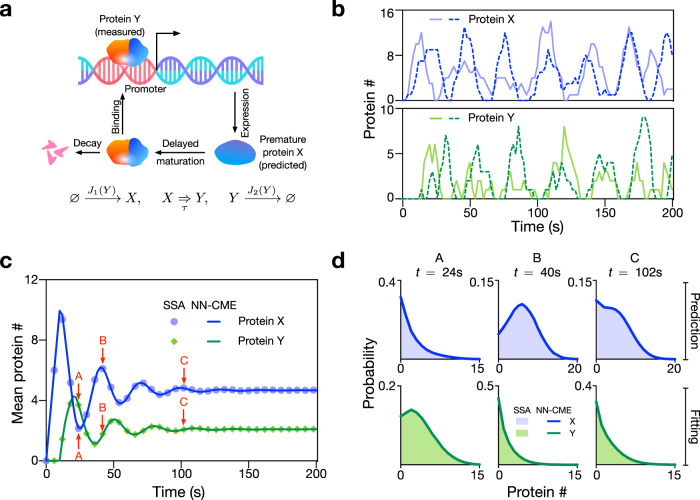
Fig. 6. NN-CME accurately predicts the properties of a stochastic auto-regulatory model of oscillatory gene expression when only partial data are used for ANN training.
a Illustration of a model of auto-regulation whereby a protein X is transcribed by a gene, then it is transformed after a delay time τ into a mature protein Y, which binds the promoter and represses transcription of X. The functions J1(Y) and J2(Y) can be found in SI Note 7. b Two typical SSA simulations of proteins X and Y, clearly showing that single-cell oscillations while noisy, they are sustained. c, d The NN-CME is obtained from training the ANN using only protein Y data from SSA simulations of the delay model of the auto-regulatory model. Surprisingly, the NN-CME’s solution for the temporal variation of the mean number of both proteins X and Y, and for their distributions is in excellent agreement with that of the SSA. Note the distributions in (d) are for the three time points labelled A, B and C in (c). The rate constants and other parameters related to the ANN’s training are specified in SI Table 1.