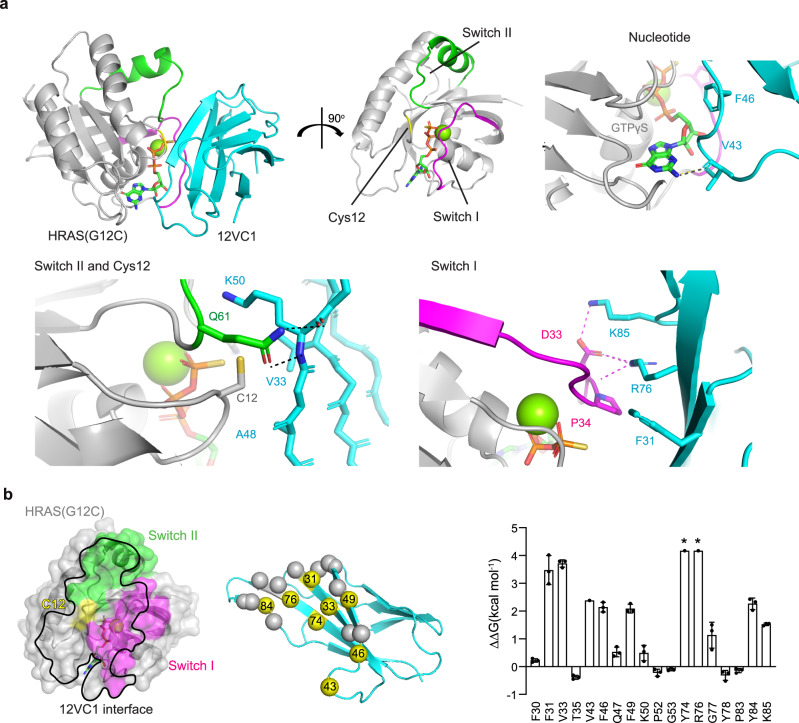
Fig. 2. Structural basis for mutant-selective recognition of RAS by 12VC1 monobody.
a Crystal structure of 12VC1 (cyan) in complex with HRAS(G12C) bound to GTPγS (gray) at 2.54 Å resolution (PDB: 7L0G). Critical interactions between 12VC1 and HRAS(G12C) occur at the switch I (magenta) and switch II (green) regions. Interactions at these regions are expanded for detailed views in separate panels. Residue V43 of 12VC1 forms a hydrogen bound with the nucleotide. Mutated position, Cys12 of RAS (yellow), is accommodated by a pocket that consists of residues V33, A48, and K50 of 12VC1 shown in stick model. Residues R76 and K85 on 12VC1 form hydrogen bond and salt bridges with Switch I residue D33. Residue F31 on 12VC1 forms hydrophobic interactions with Switch I. b An open book view of the HRAS(G12C):12VC1 complex. Effects of alanine mutation of 12VC1 residues located within 4 Å of HRAS are shown in the bar graph (mean ± SD). Asterisks denote that ΔΔG is beyond the measurable limit of experiments and thus the values shown represent the lower limit. Experiment was performed in triplicate except for V43, Y74, and R76, and the error bars indicate s.d. The mutated residues in alanine scanning are shown as spheres in the cartoon model, and those for critical residues (ΔΔG > 2 kcal/mol) are colored yellow and labeled (middle panel).