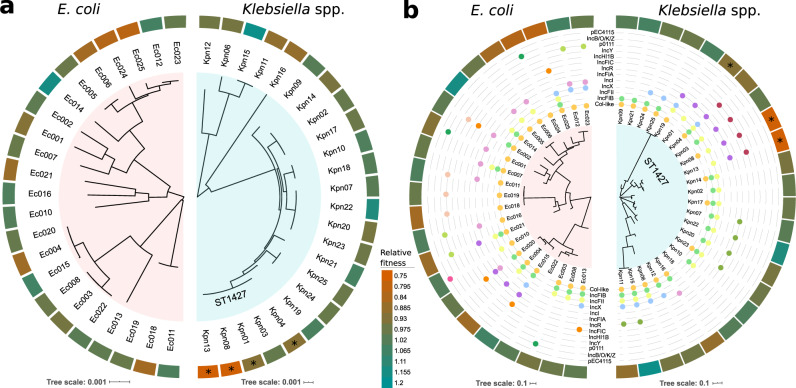
Fig. 4. Fitness effects of pOXA-48_K8 across bacterial genome content.
An association was found between pOXA-48_K8 fitness effects and bacterial host genomic content for four K. pneumoniae ST1427 isolates. a Core genome relationships among E. coli (left) and Klebsiella spp. (right). Tree construction is based on polymorphisms in the core genome. The outer circle indicates the relative fitness of pOXA-48-carrying bacterial hosts (see legend for colour code; red indicates fitness costs and green indicates fitness benefits associated with pOXA-48_K8 carriage). Asterisks denote clones with a phylogenetic signal associated with plasmid fitness effects (LIPA, P < 0.05). b Accessory genome relationships among E. coli (left) and Klebsiella spp. (right) isolates. This tree is a gene content tree constructed based on the distance matrix of the accessory gene network of each group. The outermost circle indicates relative fitness as in a. The intermediate circles indicate presence/absence of plasmids belonging to the different plasmid families named in the figure. Note that only two isolates do not carry any plasmids. Asterisks denote clones with a significant phylogenetic signal associating accessory genome composition with pOXA-48 fitness effects (LIPA, P < 0.05).