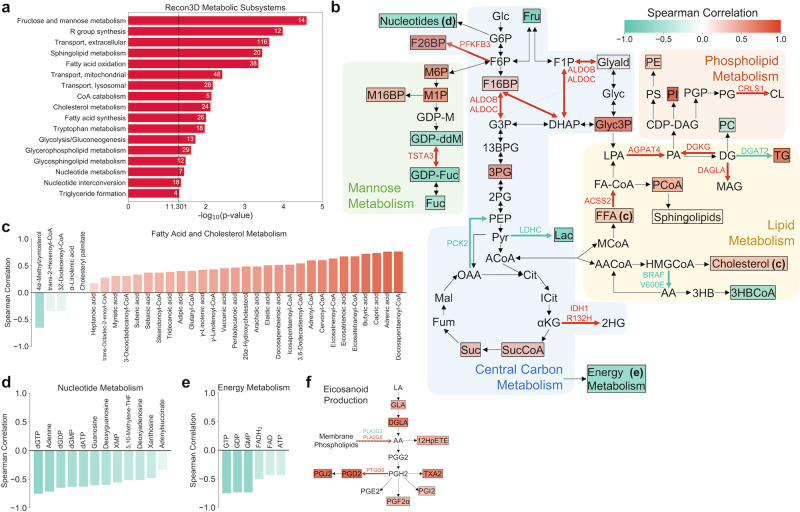
Fig. 5. Analysis of metabolic biomarkers from the multi-omics classifier for radiation response.
a Metabolite set enrichment analysis (MSEA) of significant metabolic features among Recon3D metabolic subsystems. The numbers of significant metabolites in each subsystem are shown. Only statistically significant (p ≤ 0.05) subsystems are shown. b Overview of significant metabolic features, as well as metabolism-related gene expression and mutation features. Different metabolic pathways are shown with colored backgrounds. Significant metabolic features are denoted by colored boxes, where the color indicates the Spearman correlation coefficient between SHAP value (ΔP) and predicted metabolite production rate across all patients (Supplementary Fig. 19). Significant gene expression and mutation features are denoted by colored reaction arrows, either in green (associated with radiation sensitivity) or in red (associated with radiation resistance). 13BPG 1,3-bisphosphoglycerate, 2HG 2-hydroxyglutarate, 2PG 2-phosphoglycerate, 3HB 3-hydroxybutyrate, 3HBCoA 3-hydroxybutyrl-CoA, 3PG 3-phosphoglycerate, αKG Alpha-ketoglutarate, AA Acetoacetate, AACoA Acetoacetyl-CoA, ACoA Acetyl-CoA, CDP-DAG CDP-diacylglycerol, Cit Citrate, CL Cardiolipin, DG Diacylglycerol, DHAP Dihydroxyacetone phosphate, F16BP Fructose 1,6-bisphosphate, F1P Fructose 1-phosphate, F26BP Fructose 2,6-bisphosphate, F6P Fructose 6-phosphate, FA-CoA Fatty acyl-CoA, FFA Free fatty acid, Fru Fructose, Fuc Fucose, Fum Fumarate, G3P Glyceraldehyde 3-phosphate, G6P Glucose 6-phosphate, GDP-ddM GDP-4-keto-6-deoxymannose, GDP-Fuc GDP-fucose, GDP-M GDP-mannose, Glc Glucose, Glyald Glyceraldehyde, Glyc3P Glycerol 3-phosphate, Gylc Glycerol, HMGCoA 3-hydroxy-3-methylglutaryl-CoA, ICit Isocitrate, Lac Lactate, LPA Lysophosphatidic acid, M16BP Mannose 1,6-bisphosphate, M1P Mannose 1-phosphate, M6P Mannose-6-phosphate, MAG Monoacylglycerol, Mal Malate, MCoA Malonyl-CoA, OAA Oxaloacetate, PA Phosphatidic acid, PC Phosphatidylcholine, PCoA Palmitoyl-CoA, PE Phosphatidylethanolamine, PEP Phosphoenolpyruvate, PG Phosphatidylglycerol, PGP Phosphatidylglycerol-phosphate, PI Phosphatidylinositol, PS Phosphatidylserine, Pyr Pyruvate, Suc Succinate, SucCoA Succinyl-CoA, TG Triglyceride. c–e Spearman correlation coefficients of significant metabolic features involved in c fatty acid and cholesterol metabolism, d nucleotide metabolism, and e energy metabolism. f Metabolic pathway of eicosanoid production, highlighting significant metabolite and gene expression features. 12HpETE 12-hydroxyperoxyeicosatetraenoic acid, AA Arachidonic acid, DGLA Dihomo-γ-linolenic acid, GLA γ-linolenic acid, LA linoleic acid.